Mitochondrial fusion in yeast requires the transmembrane GTPase Fzo1p
- PMID: 9786948
- PMCID: PMC2132826
- DOI: 10.1083/jcb.143.2.359
Mitochondrial fusion in yeast requires the transmembrane GTPase Fzo1p
Abstract
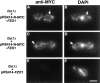
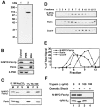
Membrane fusion is required to establish the morphology and cellular distribution of the mitochondrial compartment. In Drosophila, mutations in the fuzzy onions (fzo) GTPase block a developmentally regulated mitochondrial fusion event during spermatogenesis. Here we report that the yeast orthologue of fuzzy onions, Fzo1p, plays a direct and conserved role in mitochondrial fusion. A conditional fzo1 mutation causes the mitochondrial reticulum to fragment and blocks mitochondrial fusion during yeast mating. Fzo1p is a mitochondrial integral membrane protein with its GTPase domain exposed to the cytoplasm. Point mutations that alter conserved residues in the GTPase domain do not affect Fzo1p localization but disrupt mitochondrial fusion. Suborganellar fractionation suggests that Fzo1p spans the outer and is tightly associated with the inner mitochondrial membrane. This topology may be required to coordinate the behavior of the two mitochondrial membranes during the fusion reaction. We propose that the fuzzy onions family of transmembrane GTPases act as molecular switches to regulate a key step in mitochondrial membrane docking and/or fusion.
Figures





References
-
- Adari H, Lowy DR, Willumsen BM, Der CJ, McCormick F. Guanosine triphosphatase activating protein (GAP) interacts with the p21 raseffector binding domain. Science. 1988;240:518–521. - PubMed
-
- Bakeeva LE, Chentsov YS, Skulachev VP. Mitochondrial framework (reticulum mitochondriale) in rat diaphragm muscle. Biochim Biophys Acta. 1978;501:349–369. - PubMed
-
- Bakeeva LE, Chentsov YS, Skulachev VP. Ontogenesis of mitochondrial reticulum in rat diaphragm muscle. Eur J Cell Biol. 1981;25:175–181. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

