Identification of differentially expressed mRNA in prokaryotic organisms by customized amplification libraries (DECAL): the effect of isoniazid on gene expression in Mycobacterium tuberculosis
- PMID: 9789070
- PMCID: PMC23765
- DOI: 10.1073/pnas.95.22.13227
Identification of differentially expressed mRNA in prokaryotic organisms by customized amplification libraries (DECAL): the effect of isoniazid on gene expression in Mycobacterium tuberculosis
Abstract
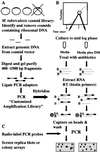
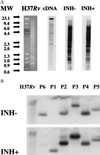
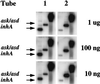
Understanding the effects of the external environment on bacterial gene expression can provide valuable insights into an array of cellular mechanisms including pathogenesis, drug resistance, and, in the case of Mycobacterium tuberculosis, latency. Because of the absence of poly(A)+ mRNA in prokaryotic organisms, studies of differential gene expression currently must be performed either with large amounts of total RNA or rely on amplification techniques that can alter the proportional representation of individual mRNA sequences. We have developed an approach to study differences in bacterial mRNA expression that enables amplification by the PCR of a complex mixture of cDNA sequences in a reproducible manner that obviates the confounding effects of selected highly expressed sequences, e.g., ribosomal RNA. Differential expression using customized amplification libraries (DECAL) uses a library of amplifiable genomic sequences to convert total cellular RNA into an amplified probe for gene expression screens. DECAL can detect 4-fold differences in the mRNA levels of rare sequences and can be performed on as little as 10 ng of total RNA. DECAL was used to investigate the in vitro effect of the antibiotic isoniazid on M. tuberculosis, and three previously uncharacterized isoniazid-induced genes, iniA, iniB, and iniC, were identified. The iniB gene has homology to cell wall proteins, and iniA contains a phosphopantetheine attachment site motif suggestive of an acyl carrier protein. The iniA gene is also induced by the antibiotic ethambutol, an agent that inhibits cell wall biosynthesis by a mechanism that is distinct from isoniazid. The DECAL method offers a powerful new tool for the study of differential gene expression.
Figures


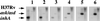
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

