Analysis of the bacteriolytic enzymes of the autolytic lactococcus lactis subsp. cremoris strain AM2 by renaturing polyacrylamide gel electrophoresis: identification of a prophage-encoded enzyme
- PMID: 9797258
- PMCID: PMC106620
- DOI: 10.1128/AEM.64.11.4142-4148.1998
Analysis of the bacteriolytic enzymes of the autolytic lactococcus lactis subsp. cremoris strain AM2 by renaturing polyacrylamide gel electrophoresis: identification of a prophage-encoded enzyme
Abstract
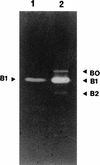
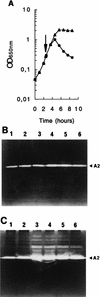
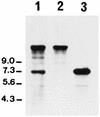
Lactococcus lactis subsp. cremoris AM2 was previously shown to lyse early and extensively during cheese ripening (M.-P. Chapot-Chartier, C. Deniel, M. Rousseau, L. Vassal, and J.-C. Gripon, Int. Dairy J. 4:251-269, 1994). We analyzed the bacteriolytic activities of autolytic strain AM2 by using renaturing sodium dodecyl sulfate-polyacrylamide gel electrophoresis performed with two different substrates in the gel, Micrococcus lysodeikticus and L. lactis autoclaved cells. Several lytic activities were detected in L. lactis AM2; a major lytic activity, designated A2 (46 kDa), was found only with the L. lactis cell substrate. This activity appears to be different from major peptidoglycan hydrolase AcmA characterized previously (G. Buist, J. Kok, K. J. Leenhouts, M. Dabrowska, G. Venema, and A. J. Haandrickman, J. Bacteriol. 177:1554-1563, 1995), which has a similar molecular mass. The two enzymes differ in substrate specificity as well as in sensitivity to pH and different chemical compounds. L. lactis AM2 is lysogenic and mitomycin C inducible. Enzyme A2 was shown to be inducible by mitomycin C and to be prophage encoded. It was identified as an enzyme similar to the lysin encoded by lactococcal small isometric temperate bacteriophages. A prophage-cured derivative of L. lactis AM2 was obtained, and this isolate exhibited different autolytic properties than AM2. After prolonged incubation in the stationary phase after growth on M17 medium, the extent of lysis of an AM2 culture was 60%, whereas over the same period there was almost no lysis in a prophage-cured derivative strain culture. These results suggest that the prophage lytic system is involved in the strain AM2 lysis observed in liquid medium and that it could also be involved in the lysis observed during cheese ripening.
Figures



References
-
- Accolas J-P, Spillmann H. The morphology of six bacteriophages of Streptococcus thermophilus. J Appl Bacteriol. 1979;47:135–144.
-
- Bie R, Sjöström G. Autolytic properties of some lactic acid bacteria used in cheese production. Part I. Material and methods. Milchwissenschaft. 1975;30:653–657.
-
- Bie R, Sjöström G. Autolytic properties of some lactic acid bacteria used in cheese production. Part II. Experiments with fluid substrates and cheese. Milchwissenschaft. 1975;30:739–747.
-
- Birkeland N-K. Cloning, molecular characterization, and expression of the genes encoding the lytic functions of lactococcal bacteriophage φLC3: a dual lysis system of modular design. Can J Microbiol. 1994;40:658–665. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

