Analysis of subfossil molecular remains of purple sulfur bacteria in a lake sediment
- PMID: 9797316
- PMCID: PMC106678
- DOI: 10.1128/AEM.64.11.4513-4521.1998
Analysis of subfossil molecular remains of purple sulfur bacteria in a lake sediment
Abstract
Molecular remains of purple sulfur bacteria (Chromatiaceae) were detected in Holocene sediment layers of a meromictic salt lake (Mahoney Lake, British Columbia, Canada). The carotenoid okenone and bacteriophaeophytin a were present in sediments up to 11,000 years old. Okenone is specific for only a few species of Chromatiaceae, including Amoebobacter purpureus, which presently predominates in the chemocline bacterial community of the lake. With a primer set specific for Chromatiaceae in combination with denaturing gradient gel electrophoresis, 16S rRNA gene sequences of four different Chromatiaceae species were retrieved from different depths of the sediment. One of the sequences, which originated from a 9, 100-year-old sample, was 99.2% identical to the 16S rRNA gene sequence of A. purpureus ML1 isolated from the chemocline. Employing primers specific for A. purpureus ML1 and dot blot hybridization of the PCR products, the detection limit for A. purpureus ML1 DNA could be lowered to 0.004% of the total community DNA. With this approach the DNA of the isolate was detected in 7 of 10 sediment layers, indicating that A. purpureus ML1 constituted at least a part of the ancient purple sulfur bacterial community. The concentrations of A. purpureus DNA and okenone in the sediment were not correlated, and the ratio of DNA to okenone was much lower in the subfossil sediment layers (2.7 . 10(-6)) than in intact cells (1.4). This indicates that degradation rates are significantly higher for genomic DNA than for hydrocarbon cell constituents, even under anoxic conditions and at the very high sulfide concentrations present in Mahoney Lake.
Figures



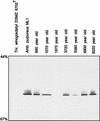


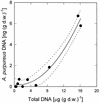

References
-
- Brown S R, McIntosh H J. The fossil history of sulfur phototrophs in a meromictic lake ecosystem. Acta Academica Aboensis. 1987;47:83–95.
-
- Brown S R, McIntosh H J, Smol J P. Recent paleolimnology of a meromictic lake: fossil pigments of photosynthetic bacteria. Verh Int Ver Limnol. 1984;22:1357–1360.
-
- Carpenter S R, Elser M M, Elser J J. Chlorophyll production, degradation, and sedimentation: implications for paleolimnology. Limnol Oceanogr. 1986;31:112–124.
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

