Kinetic bias in estimates of coastal picoplankton community structure obtained by measurements of small-subunit rRNA gene PCR amplicon length heterogeneity
- PMID: 9797317
- PMCID: PMC106679
- DOI: 10.1128/AEM.64.11.4522-4529.1998
Kinetic bias in estimates of coastal picoplankton community structure obtained by measurements of small-subunit rRNA gene PCR amplicon length heterogeneity
Abstract
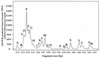
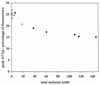
Marine bacterioplankton diversity was examined by quantifying natural length variation in the 5' domain of small-subunit (SSU) rRNA genes (rDNA) amplified by PCR from a DNA sample from the Oregon coast. This new technique, length heterogeneity analysis by PCR (LH-PCR), determines the relative proportions of amplicons originating from different organisms by measuring the fluorescence emission of a labeled primer used in the amplification reaction. Relationships between the sizes of amplicons and gene phylogeny were predicted by an analysis of 366 SSU rDNA sequences from cultivated marine bacteria and from bacterial genes cloned directly from environmental samples. LH-PCR was used to compare the distribution of bacterioplankton SSU rDNAs from a coastal water sample with that of an SSU rDNA clone library prepared from the same sample and also to examine the distribution of genes in the PCR products from which the clone library was prepared. The analysis revealed that the relative frequencies of genes amplified from natural communities are highly reproducible for replicate sets of PCRs but that a bias possibly caused by the reannealing kinetics of product molecules can skew gene frequencies when PCR product concentrations exceed threshold values.
Figures

References
-
- Giovannoni S J. The polymerase chain reaction. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. New York, N.Y: John Wiley & Sons, Inc.; 1991. pp. 177–203.
-
- Giovannoni S J, Britschgi T B, Moyer C L, Field K G. Genetic diversity in Sargasso Sea bacterioplankton. Nature. 1990;345:60–63. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

