Quantification of 16S rRNAs in complex bacterial communities by multiple competitive reverse transcription-PCR in temperature gradient gel electrophoresis fingerprints
- PMID: 9797325
- PMCID: PMC106687
- DOI: 10.1128/AEM.64.11.4581-4587.1998
Quantification of 16S rRNAs in complex bacterial communities by multiple competitive reverse transcription-PCR in temperature gradient gel electrophoresis fingerprints
Abstract
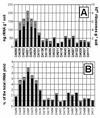
A novel approach was developed to quantify rRNA sequences in complex bacterial communities. The main bacterial 16S rRNAs in Drentse A grassland soils (The Netherlands) were amplified by reverse transcription (RT)-PCR with bacterium-specific primers and were separated by temperature gradient gel electrophoresis (TGGE). The primer pair used (primers U968-GC and L1401) was found to amplify with the same efficiency 16S rRNAs from bacterial cultures containing different taxa and cloned 16S ribosomal DNA amplicons from uncultured soil bacteria. The sequence-specific efficiency of amplification was determined by monitoring the amplification kinetics by kinetic PCR. The primer-specific amplification efficiency was assessed by competitive PCR and RT-PCR, and identical input amounts of different 16S rRNAs resulted in identical amplicon yields. The sequence-specific detection system used for competitive amplifications was TGGE, which also has been found to be suitable for simultaneous quantification of more than one sequence. We demonstrate that this approach can be applied to TGGE fingerprints of soil bacteria to estimate the ratios of the bacterial 16S rRNAs.
Figures
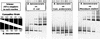




References
-
- Alard P, Lantz O, Sebagh M, Calvo C F, Weill D, Chavanel G, Senik A, Charpentier B. A versatile ELISA-PCR assay for mRNA quantitation from a few cells. BioTechniques. 1993;15:730–737. - PubMed
-
- Blok H J, Gohlke A, Akkermans A D L. Quantitative analysis of 16S rDNA using competitive PCR and the QPCR System 5000. BioTechniques. 1997;22:700–704. - PubMed
-
- Buck K J, Harris R A, Sikela J M. A general method for quantitative PCR analysis of mRNA levels for members of gene families: application to GABAA receptor subunits. BioTechniques. 1991;11:636–641. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

