Regulation of the anaphase-promoting complex/cyclosome by bimAAPC3 and proteolysis of NIMA
- PMID: 9802893
- PMCID: PMC25582
- DOI: 10.1091/mbc.9.11.3019
Regulation of the anaphase-promoting complex/cyclosome by bimAAPC3 and proteolysis of NIMA
Abstract
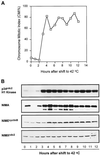
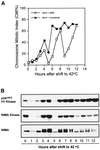
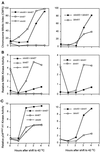
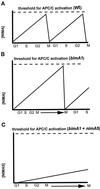
Surprisingly, although highly temperature-sensitive, the bimA1(APC3) anaphase-promoting complex/cyclosome (APC/C) mutation does not cause arrest of mitotic exit. Instead, rapid inactivation of bimA1(APC3) is shown to promote repeating oscillations of chromosome condensation and decondensation, activation and inactivation of NIMA and p34(cdc2) kinases, and accumulation and degradation of NIMA, which all coordinately cycle multiple times without causing nuclear division. These bimA1(APC3)-induced cell cycle oscillations require active NIMA, because a nimA5 + bimA1(APC3) double mutant arrests in a mitotic state with very high p34(cdc2) H1 kinase activity. NIMA protein instability during S phase and G2 was also found to be controlled by the APC/C. The bimA1(APC3) mutation therefore first inactivates the APC/C but then allows its activation in a cyclic manner; these cycles depend on NIMA. We hypothesize that bimAAPC3 could be part of a cell cycle clock mechanism that is reset after inactivation of bimA1(APC3). The bimA1(APC3) mutation may also make the APC/C resistant to activation by mitotic substrates of the APC/C, such as cyclin B, Polo, and NIMA, causing mitotic delay. Once these regulators accumulate, they activate the APC/C, and cells exit from mitosis, which then allows this cycle to repeat. The data indicate that bimAAPC3 regulates the APC/C in a NIMA-dependent manner.
Figures




Similar articles
-
BIMAAPC3, a component of the Aspergillus anaphase promoting complex/cyclosome, is required for a G2 checkpoint blocking entry into mitosis in the absence of NIMA function.J Cell Sci. 1998 May;111 ( Pt 10):1453-65. doi: 10.1242/jcs.111.10.1453. J Cell Sci. 1998. PMID: 9570762
-
Xe-p9, a Xenopus Suc1/Cks protein, is essential for the Cdc2-dependent phosphorylation of the anaphase- promoting complex at mitosis.Genes Dev. 1998 Aug 15;12(16):2549-59. doi: 10.1101/gad.12.16.2549. Genes Dev. 1998. PMID: 9716407 Free PMC article.
-
Chromosome separation and exit from mitosis in budding yeast: dependence on growth revealed by cAMP-mediated inhibition.Exp Cell Res. 1999 Aug 1;250(2):510-23. doi: 10.1006/excr.1999.4531. Exp Cell Res. 1999. PMID: 10413604
-
Regulation of p34cdc2/cyclinB H1 and NIMA kinases during the G2/M transition and checkpoint responses in Aspergillus nidulans.Prog Cell Cycle Res. 1997;3:221-32. doi: 10.1007/978-1-4615-5371-7_17. Prog Cell Cycle Res. 1997. PMID: 9552417 Review.
-
Subunits and substrates of the anaphase-promoting complex.Exp Cell Res. 1999 May 1;248(2):339-49. doi: 10.1006/excr.1999.4443. Exp Cell Res. 1999. PMID: 10222126 Review.
Cited by
-
Cell cycle progression and cell polarity require sphingolipid biosynthesis in Aspergillus nidulans.Mol Cell Biol. 2001 Sep;21(18):6198-209. doi: 10.1128/MCB.21.18.6198-6209.2001. Mol Cell Biol. 2001. PMID: 11509663 Free PMC article.
-
Hypomorphic bimA(APC3) alleles cause errors in chromosome metabolism that activate the DNA damage checkpoint blocking cytokinesis in Aspergillus nidulans.Genetics. 2000 Jan;154(1):167-79. doi: 10.1093/genetics/154.1.167. Genetics. 2000. PMID: 10628978 Free PMC article.
-
Modeling the fission yeast cell cycle: quantized cycle times in wee1- cdc25Delta mutant cells.Proc Natl Acad Sci U S A. 2000 Jul 5;97(14):7865-70. doi: 10.1073/pnas.97.14.7865. Proc Natl Acad Sci U S A. 2000. PMID: 10884416 Free PMC article.
-
Insights into dynamic mitotic chromatin organization through the NIMA kinase suppressor SonC, a chromatin-associated protein involved in the DNA damage response.Genetics. 2014 Jan;196(1):177-95. doi: 10.1534/genetics.113.156745. Epub 2013 Nov 8. Genetics. 2014. PMID: 24214344 Free PMC article.
-
Gamma-tubulin regulates the anaphase-promoting complex/cyclosome during interphase.J Cell Biol. 2010 Aug 9;190(3):317-30. doi: 10.1083/jcb.201002105. Epub 2010 Aug 2. J Cell Biol. 2010. PMID: 20679430 Free PMC article.
References
-
- Alfa CE, Ducommun B, Beach D, Hyams JS. Distinct nuclear and spindle pole body populations of cyclin-cdc2 in fission yeast. Nature. 1990;347:680–682. - PubMed
-
- Amon A, Irniger S, Nasmyth K. Closing the cell cycle circle in yeast: G2 cyclin proteolysis initiated at mitosis persists until the activation of G1 cyclins in the next cycle. Cell. 1994;77:1037–1050. - PubMed
-
- Charles JF, Jaspersen SL, Tinker-Kulberg RL, Hwang L, Szidon A, Morgan DO. The polo-related kinase cdc5 activates and is destroyed by the mitotic cyclin destruction machinery in S. cerevisiae. Curr Biol. 1998;8:497–507. - PubMed
-
- Cohen-Fix O, Koshland D. The metaphase-to-anaphase transition: avoiding a mid-life crisis. Curr Opin Cell Biol. 1997;9:800–806. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

