tRNA nuclear export in saccharomyces cerevisiae: in situ hybridization analysis
- PMID: 9802895
- PMCID: PMC25586
- DOI: 10.1091/mbc.9.11.3041
tRNA nuclear export in saccharomyces cerevisiae: in situ hybridization analysis
Abstract
To understand the factors specifically affecting tRNA nuclear export, we adapted in situ hybridization procedures to locate endogenous levels of individual tRNA families in wild-type and mutant yeast cells. Our studies of tRNAs encoded by genes lacking introns show that nucleoporin Nup116p affects both poly(A) RNA and tRNA export, whereas Nup159p affects only poly(A) RNA export. Los1p is similar to exportin-t, which facilitates vertebrate tRNA export. A los1 deletion mutation affects tRNA but not poly(A) RNA export. The data support the notion that Los1p and exportin-t are functional homologues. Because LOS1 is nonessential, tRNA export in vertebrate and yeast cells likely involves factors in addition to exportin-t. Mutation of RNA1, which encodes RanGAP, causes nuclear accumulation of tRNAs and poly(A) RNA. Many yeast mutants, including those with the rna1-1 mutation, affect both pre-tRNA splicing and RNA export. Our studies of the location of intron-containing pre-tRNAs in the rna1-1 mutant rule out the possibility that this results from tRNA export occurring before splicing. Our results also argue against inappropriate subnuclear compartmentalization causing defects in pre-tRNA splicing. Rather, the data support "feedback" of nucleus/cytosol exchange to the pre-tRNA splicing machinery.
Figures

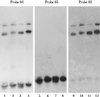
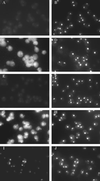

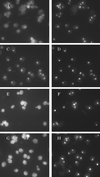

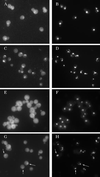
References
-
- Amberg DC, Goldstein AL, Cole CN. Isolation and characterization of RAT1: an essential gene of Saccharomyces cerevisiae required for the efficient nucleocytoplasmic trafficking of mRNA. Genes Dev. 1992;6:1173–1189. - PubMed
-
- Arts G, Fornerod M, Mattaj IW. Identification of a nuclear export receptor for tRNA. Curr Biol. 1998;8:305–314. - PubMed
-
- Becker J, Melchior F, Gerke V, Bischoff FR, Ponstingl H, Wittinghofer A. RNA1 encodes a GTPase-activating protein specific for Gsp1p, the Ran/TC4 homologue of Saccharomyces cerevisiae. J Biol Chem. 1995;270:11860–11865. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

