Characterization of gene expression in resting and activated mast cells
- PMID: 9802978
- PMCID: PMC2212524
- DOI: 10.1084/jem.188.9.1657
Characterization of gene expression in resting and activated mast cells
Erratum in
- J Exp Med 1998 Dec 21;188(12):2387
Abstract
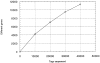
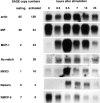
To characterize gene expression in activated mast cells more comprehensively than heretofore, we surveyed the changes in genetic transcripts by the method of serial analysis of gene expression in the RBL-2H3 line of rat mast cells before and after they were stimulated through their receptors with high affinity for immunoglobulin E (FcepsilonRI). A total of 40,759 transcripts derived from 11,300 genes were analyzed. Among the diverse genes that had not been previously associated with mast cells and that were constitutively expressed were those for the cytokine macrophage migration inhibitory factor neurohormone receptors such as growth hormone- releasing factor and melatonin and components of the exocytotic machinery. In addition, several dozen transcripts were differentially expressed in response to antigen-induced clustering of the FcepsilonRI. Included among these were the genes for preprorelaxin, mitogen-activated protein kinase kinase 3, and the dual specificity protein phosphatase, rVH6. Significantly, the majority of genes differentially expressed in this well-studied model of mast cell activation have not been identified before this analysis.
Figures



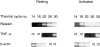
References
-
- Selye, H. 1965. The Mast Cells. Butterworth Inc., Washington DC. 1–498.
-
- Rigby, L., M.D. Hulett, R.I. Brinkworth, and P.M. Hogarth. 1998. The structural basis of the interaction of IgE and FcεRI. In IgE Receptor (FcεRI) Function in Mast Cells and Basophils. M.M. Hamawy, editor. R.G. Landes Company, Austin, TX. 7–32.
-
- Hamawy, M.M., and W.D. Swaim. 1997. FcεRI-mediated cell degranulation, proliferation and adhesion. In IgE Receptor (FcεRI) Function in Mast Cells and Basophils. M.M. Hamawy, editor. R.G. Landes Company, Austin, TX. 173–180.
-
- Gordon, J.R. 1997. FcεRI-induced cytokine production and gene expression. In IgE Receptor (FcεRI) Function in Mast Cells and Basophils. M.M. Hamawy, editor. R.G. Landes Company, Austin, TX. 209–242.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

