Acid-growth response and alpha-expansins in suspension cultures of bright yellow 2 tobacco
- PMID: 9808735
- PMCID: PMC34801
- DOI: 10.1104/pp.118.3.907
Acid-growth response and alpha-expansins in suspension cultures of bright yellow 2 tobacco
Abstract
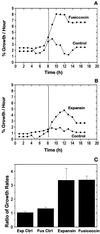
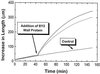
The possibility that Bright Yellow 2 (BY2) tobacco (Nicotiana tabacum L.) suspension-cultured cells possess an expansin-mediated acid-growth mechanism was examined by multiple approaches. BY2 cells grew three times faster upon treatment with fusicoccin, which induces an acidification of the cell wall. Exogenous expansins likewise stimulated BY2 cell growth 3-fold. Protein extracted from BY2 cell walls possessed the expansin-like ability to induce extension of isolated walls. In western-blot analysis of BY2 wall protein, one band of 29 kD was recognized by anti-expansin antibody. Six different classes of alpha-expansin mRNA were identified in a BY2 cDNA library. Northern-blot analysis indicated moderate to low abundance of multiple alpha-expansin mRNAs in BY2 cells. From these results we conclude that BY2 suspension-cultured cells have the necessary components for expansin-mediated cell wall enlargement.
Figures





References
-
- Akehurst BC (1970) The botany, genetics and the development of commercial types. In D Rhind, ed, Tobacco, Ed 2. Longham Group, London, pp 42–61
-
- Cabot EL, Beckenbach AT. Simultaneous editing of multiple nucleic acid and protein sequences with ESEE. Comput Appl Biosci. 1989;5:233–234. - PubMed
-
- Carpita NC, Gibeaut DM. Structural models of primary cell walls in flowering plants: consistency of molecular structure with the physical properties of the walls during growth. Plant J. 1993;3:1–30. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

