Autolysis of Lactococcus lactis is influenced by proteolysis
- PMID: 9811653
- PMCID: PMC107669
- DOI: 10.1128/JB.180.22.5947-5953.1998
Autolysis of Lactococcus lactis is influenced by proteolysis
Abstract
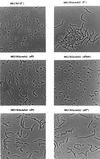
The autolysin AcmA of Lactococcus lactis was shown to be degraded by the extracellular lactococcal proteinase PrtP. Autolysis, as evidenced by reduction in optical density of a stationary-phase culture and concomitant release of intracellular proteins, was greatly reduced when L. lactis MG1363 cells expressed the cell wall-anchored lactococcal proteinase PrtP of the PI-type caseinolytic specificity (PI). On the other hand, lactococcal strains that did not produce the proteinase showed a high level of autolysis, which was also observed when the cells produced the secreted form of PI or a cell wall-anchored proteinase with PIII-type specificity. Autolysis was also increased when MG1363 expressed the cell wall-anchored hybrid PI/PIII-type proteinase PIac. Zymographic analysis of AcmA activity during stationary phase showed that AcmA was quickly degraded by PI and much more slowly by PrtP proteinases with PIII-type and intermediate specificities. Autolysis of L. lactis by AcmA was influenced by the specificity, amount, and location of the lactococcal proteinase. No autolysis was observed when the various proteinases were expressed in an L. lactis acmA deletion mutant, indicating that PrtP itself did not cause lysis of cells. The chain length of a strain was significantly shortened when the strain expressed a cell wall-anchored active proteinase.
Figures



References
-
- Buist G. AcmA of Lactococcus lactis, a cell-binding muramidase. Ph.D. thesis. Groningen, The Netherlands: University of Groningen; 1997.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

