The ectodomain of a novel member of the immunoglobulin subfamily related to the poliovirus receptor has the attributes of a bona fide receptor for herpes simplex virus types 1 and 2 in human cells
- PMID: 9811737
- PMCID: PMC110516
- DOI: 10.1128/JVI.72.12.9992-10002.1998
The ectodomain of a novel member of the immunoglobulin subfamily related to the poliovirus receptor has the attributes of a bona fide receptor for herpes simplex virus types 1 and 2 in human cells
Abstract
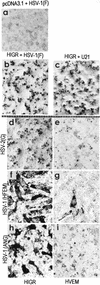
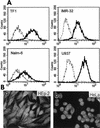
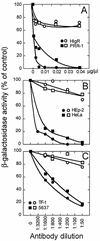
We report on the functional cloning of a hitherto unknown member of the immunoglobulin (Ig) superfamily selected for its ability to confer susceptibility to herpes simplex virus (HSV) infection on a highly resistant cell line (J1.1-2 cells), derived by exposure of BHKtk- cells to a recombinant HSV-1 expressing tumor necrosis factor alpha (TNF-alpha). The sequence of herpesvirus Ig-like receptor (HIgR) predicts a transmembrane protein with an ectodomain consisting of three cysteine-bracketed domains, one V-like and two C-like. HIgR shares its ectodomain with and appears to be an alternative splice variant of the previously described protein PRR-1 (poliovirus receptor-related protein). Both HIgR and PRR-1 conferred on J1.1-2 cells susceptibility to HSV-1, HSV-2, and bovine herpesvirus 1. The viral ligand of HIgR and PRR-1 is glycoprotein D, a constituent of the virion envelope long known to mediate viral entry into cells through interaction with cellular receptor molecules. Recently, PRR-1, renamed HveC (herpesvirus entry mediator C), and the related PRR-2, renamed HveB, were reported to mediate the entry of HSV-1, HSV-2, and bovine herpesvirus 1, and the homologous poliovirus receptor was reported to mediate the entry of pseudorabies virus (R. J. Geraghty, C. Krummenacher, G. H. Cohen, R. J. Eisenberg, and P. G. Spear, Science 280:1618-1620, 1998; M. S. Warner, R. J. Geraghty, W. M. Martinez, R. I. Montgomery, J. C. Whitbeck, R. Xu, R. J. Eisenberg, G. H. Cohen, and P. G. Spear, Virology 246:179-189, 1998). Here we further show that HIgR or PRR-1 proteins detected by using a monoclonal antibody to PRR-1 are widely distributed among human cell lines susceptible to HSV infection and commonly used for HSV studies. The monoclonal antibody neutralized virion infectivity in cells transfected with HIgR or PRR-1 cDNA, as well as in the human cell lines, indicating a direct interaction of virions with the receptor molecule, and preliminarily mapping this function to the ectodomain of HIgR and PRR-1. Northern blot analysis showed that HIgR or PRR-1 mRNAs were expressed in human tissues, with the highest expression being detected in nervous system samples. HIgR adds a novel member to the cluster of Ig superfamily members able to mediate the entry of alphaherpesviruses into cells. The wide distribution of HIgR or PRR-1 proteins among human cell lines susceptible to HSV infection, coupled with the neutralizing activity of the antibody in the same cells, provides direct demonstration of the actual use of this cluster of molecules as HSV-1 and HSV-2 entry receptors in human cell lines. The high level of expression in samples from nervous system makes the use of these proteins in human tissues very likely. This cluster of molecules may therefore be considered to constitute bona fide receptors for HSV-1 and HSV-2.
Figures

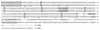



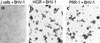

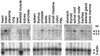
References
-
- Bonaldo M F, Lennon G, Soares M B. Normalization and subtraction: two approaches to facilitate gene discovery. Genome Res. 1996;6:791–806. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

