Simian T-cell lymphotropic virus type 1 from Mandrillus sphinx as a simian counterpart of human T-cell lymphotropic virus type 1 subtype D
- PMID: 9811783
- PMCID: PMC110621
- DOI: 10.1128/JVI.72.12.10316-10322.1998
Simian T-cell lymphotropic virus type 1 from Mandrillus sphinx as a simian counterpart of human T-cell lymphotropic virus type 1 subtype D
Abstract
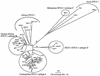
A recent serological and molecular survey of a semifree-ranging colony of mandrills (Mandrillus sphinx) living in Gabon, central Africa, indicated that 6 of 102 animals, all males, were infected with simian T-cell lymphotropic virus type 1 (STLV-1). These animals naturally live in the same forest area as do human inhabitants (mostly Pygmies) who are infected by the recently described human T-cell lymphotropic virus type 1 (HTLV-1) subtype D. We therefore investigated whether these mandrills were infected with an STLV-1 related to HTLV-1 subtype D. Nucleotide and/or amino acid sequence analyses of complete or partial long terminal repeat (LTR), env, and rex regions showed that HTLV-1 subtype D-specific mutations were found in three of four STLV-1-infected mandrills, while the remaining monkey was infected by a different STLV-1 subtype. Phylogenetic studies conducted on the LTR as well as on the env gp21 region showed that these three new STLV-1 strains from mandrills fall in the same monophyletic clade, supported by high bootstrap values, as do the sequences of HTLV-1 subtype D. These data show, for the first time, the presence of the same subtype of primate T-cell lymphotropic virus type 1 in humans and wild-caught monkeys originating from the same geographical area. This strongly supports the hypothesis that mandrills are the natural reservoir of HTLV-1 subtype D, although the possibility that another monkey species living in the same area could be the original reservoir of both human and mandrill viruses cannot be excluded. Due to the quasi-identity of both human and monkey viruses, interspecies transmission episodes leading to such a clade may have occurred recently.
Figures




References
-
- Chen J, Zekeng L, Yamashita M, Takehisa J, Miura T, Ido E, Mboudjeka I, Tsague J M, Hayami M, Kaptue L. HTLV type I isolated from a Pygmy in Cameroon is related to but distinct from the known Central African type. AIDS Res Hum Retroviruses. 1995;11:1529–1531. - PubMed
-
- Felsenstein J. PHYLIP: phylogenetic inference package, version 3.5. Seattle: University of Washington; 1993.
-
- Fultz P, Su L, May P, West J T. Isolation of sooty mangabey simian T-cell leukemia virus type 1 [STLV-1(sm)] and characterization of a mangabey T cell co-infected with STLV-1(sm) and simian immunodeficiency virus SIVPBj14. Virology. 1997;235:271–285. - PubMed
-
- Genetics Computer Group. Program manual for the GCG package, version 8.0. University of Wisconsin, Madison; 1996.
-
- Georges-Courbot M C, Moisson P, Leroy E, Pingard A M, Nerrienet E, Dubreuil G, Wickings E J, Debels F, Bedjabaga I, Poaty-Mavoungou V, Hahn N T, Georges A J. Occurrence and frequency of transmission of naturally occurring simian retroviral infections (SIV, STLV, and SRV) at the CIRMF Primate Center, Gabon. J Med Primatol. 1996;25:313–326. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

