Origins and antiquity of X-linked triallelic color vision systems in New World monkeys
- PMID: 9811872
- PMCID: PMC24891
- DOI: 10.1073/pnas.95.23.13749
Origins and antiquity of X-linked triallelic color vision systems in New World monkeys
Abstract
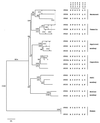
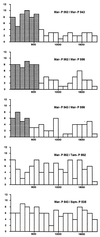
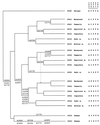
It is known that the squirrel monkey, marmoset, and other related New World (NW) monkeys possess three high-frequency alleles at the single X-linked photopigment locus, and that the spectral sensitivity peaks of these alleles are within those delimited by the human red and green pigment genes. The three alleles in the squirrel monkey and marmoset have been sequenced previously. In this study, the three alleles were found and sequenced in the saki monkey, capuchin, and tamarin. Although the capuchin and tamarin belong to the same family as the squirrel monkey and marmoset, the saki monkey belongs to a different family and is one of the species that is most divergent from the squirrel monkey and marmoset, suggesting the presence of the triallelic system in many NW monkeys. The nucleotide sequences of these alleles from the five species studied indicate that gene conversion occurs frequently and has partially or completely homogenized intronic and exonic regions of the alleles in each species, making it appear that a triallelic system arose independently in each of the five species studied. Nevertheless, a detailed analysis suggests that the triallelic system arose only once in the NW monkey lineage, from a middle wavelength (green) opsin gene, and that the amino acid differences at functionally critical sites among alleles have been maintained by natural selection in NW monkeys for >20 million years. Moreover, the two X-linked opsin genes of howler monkeys (a NW monkey genus) were evidently derived from the incorporation of a middle (green) and a long wavelength (red) allele into one chromosome; these two genes together with the (autosomal) blue opsin gene would immediately enable even a male monkey to have trichromatic vision.
Figures


References
-
- Jacobs G H, Neitz M, Deegan J F, Neitz J. Nature (London) 1996;382:156–158. - PubMed
-
- Boissinot S, Zhou Y-H, Qiu L, Dulai K S, Neiswanger K, Schneider H, Sampaio I, Hunt D M, Hewett-Emmett D, Li W-H. Zool Stud. 1997;36:360–369.
-
- Hunt D, Dulai K S, Cowing J A, Mollon J D, Bowmaker J K, Lee B B, Hewett-Emmett D, Li W-H. Vision Res. 1998;38:3299–3306. - PubMed
-
- Jacobs G H. Vision Res. 1984;24:1267–1277. - PubMed
-
- Mollon J D, Bowmaker J K, Jacobs G H. Proc R Soc London Ser B. 1984;222:373–399. - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

