Rump white inversion in the mouse disrupts dipeptidyl aminopeptidase-like protein 6 and causes dysregulation of Kit expression
- PMID: 9811881
- PMCID: PMC24902
- DOI: 10.1073/pnas.95.23.13800
Rump white inversion in the mouse disrupts dipeptidyl aminopeptidase-like protein 6 and causes dysregulation of Kit expression
Abstract
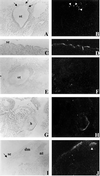
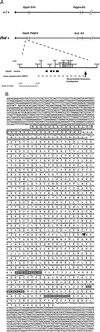
The mouse rump white (Rw) mutation causes a pigmentation defect in heterozygotes and embryonic lethality in homozygotes. At embryonic day (E) 7.5, Rw/Rw embryos are retarded in growth, fail to complete neurulation and die around E 9.5. The Rw mutation is associated with a chromosomal inversion spanning 30 cM of the proximal portion of mouse chromosome 5. The Rw embryonic lethality is complemented by the W19H deletion, which spans the distal boundary of the Rw inversion, suggesting that the Rw lethality is not caused by the disruption of a gene at the distal end of the inversion. Here, we report the molecular characterization of sequences disrupted by both inversion breakpoints. These studies indicate that the distal breakpoint of the inversion is associated with ectopic Kit expression and therefore may be responsible for the dominant pigmentation defect in Rw/+ mice; whereas the recessive lethality of Rw is probably due to the disruption of the gene encoding dipeptidyl aminopeptidase-like protein 6, Dpp6 [Wada, K., Yokotani, N., Hunter, C., Doi, K., Wenthold, R. J. & Shimasaki, S. (1992) Proc. Natl. Acad. Sci. USA 89, 197-201] located at the proximal inversion breakpoint.
Figures


References
-
- Russell W L. Cold Spring Harbor Symp Quant Biol. 1951;16:327–336. - PubMed
-
- Jackson I J. Annu Rev Genet. 1994;28:189–217. - PubMed
-
- Barsh G S. Trends Genet. 1996;12:299–305. - PubMed
-
- Searle A G, Truslove G M. Genet Res. 1970;15:227–235. - PubMed
-
- Chabot B, Stephenson D A, Chapman V M, Besmer P, Bernstein A. Nature (London) 1988;335:88–89. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

