Structure of the haemagglutinin-esterase-fusion glycoprotein of influenza C virus
- PMID: 9817207
- PMCID: PMC7095117
- DOI: 10.1038/23974
Structure of the haemagglutinin-esterase-fusion glycoprotein of influenza C virus
Abstract
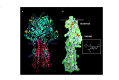
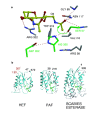
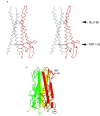
The spike glycoproteins of the lipid-enveloped orthomyxoviruses and paramyxoviruses have three functions: to recognize the receptor on the cell surface, to mediate viral fusion with the cell membrane, and to destroy the receptor. In influenza C virus, a single glycoprotein, the haemagglutinin-esterase-fusion (HEF) protein, possesses all three functions. In influenza A and B, the first two activities are mediated by haemagglutinin and the third by a second glycoprotein, neuraminidase. Here we report the crystal structure of the HEF envelope glycoprotein of influenza C virus. We have identified the receptor-binding site and the receptor-destroying enzyme (9-O-acetylesterase) sites, by using receptor analogues. The receptor-binding domain is structurally similar to the sialic acid-binding domain of influenza A haemagglutinin, but binds 9-O-acetylsialic acid. The esterase domain has a structure similar to the esterase from Streptomyces scabies and a brain acetylhydrolase. The receptor domain is inserted into a surface loop of the esterase domain and the esterase domain is inserted into a surface loop of the stem. The stem domain is similar to that of influenza A haemagglutinin, except that the triple-stranded, alpha-helical bundle diverges at both of its ends, and the amino terminus of HEF2, the fusion peptide, is partially exposed. The segregation of HEF's three functions into structurally distinct domains suggests that the entire stem region, including sequences at the amino and carboxy termini of HEF1 which precede the post-translational cleavage site between HEF1 and HEF2, forms an independent fusion domain which is probably derived from an ancestral membrane fusion protein.
Figures


References
-
- Rosenthal PB. Crystallographic Studies of the Influenza C Virus Surface Glycoprotein. 1996.
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

