Nuclear import and the evolution of a multifunctional RNA-binding protein
- PMID: 9817748
- PMCID: PMC2132966
- DOI: 10.1083/jcb.143.4.887
Nuclear import and the evolution of a multifunctional RNA-binding protein
Abstract
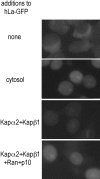
La (SS-B) is a highly expressed protein that is able to bind 3'-oligouridylate and other common RNA sequence/structural motifs. By virtue of these interactions, La is present in a myriad of nuclear and cytoplasmic ribonucleoprotein complexes in vivo where it may function as an RNA-folding protein or RNA chaperone. We have recently characterized the nuclear import pathway of the S. cerevisiae La, Lhp1p. The soluble transport factor, or karyopherin, that mediates the import of Lhp1p is Kap108p/Sxm1p. We have now determined a 113-amino acid domain of Lhp1p that is brought to the nucleus by Kap108p. Unexpectedly, this domain does not coincide with the previously identified nuclear localization signal of human La. Furthermore, when expressed in Saccharomyces cerevisiae, the nuclear localization of Schizosaccharomyces pombe, Drosophila, and human La proteins are independent of Kap108p. We have been able to reconstitute the nuclear import of human La into permeabilized HeLa cells using the recombinant human factors karyopherin alpha2, karyopherin beta1, Ran, and p10. As such, the yeast and human La proteins are imported using different sequence motifs and dissimilar karyopherins. Our results are consistent with an intermingling of the nuclear import and evolution of La.
Figures







References
-
- Aitchison JD, Blobel G, Rout MP. Kap104p: a karyopherin involved in the nuclear transport of messenger RNA binding proteins. Science. 1996;274:624–627. - PubMed
-
- Ausubel, F.M., R. Brent, R.E. Kingston, D.D. Moore, J.A. Smith, J.G. Seidman, and K. Struhl. 1997. Current Protocols in Molecular Biology. Vol. 2. John Wiley & Sons, Inc., New York. 13.0.1–13.13.9.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

