Protein kinase A-dependent derepression of the human prodynorphin gene via differential binding to an intragenic silencer element
- PMID: 9819380
- PMCID: PMC109275
- DOI: 10.1128/MCB.18.12.6921
Protein kinase A-dependent derepression of the human prodynorphin gene via differential binding to an intragenic silencer element
Abstract
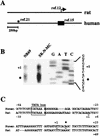
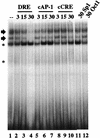
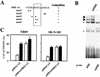
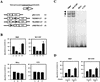
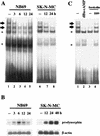
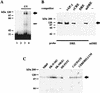
Induction of the prodynorphin gene has been implicated in medium and long-term adaptation during memory acquisition and pain. By 5' deletion mapping and site-directed mutagenesis of the human prodynorphin promoter, we demonstrate that both basal transcription and protein kinase A (PKA)-induced transcription in NB69 and SK-N-MC human neuroblastoma cells are regulated by the GAGTCAAGG sequence centered at position +40 in the 5' untranslated region of the gene (named the DRE, for downstream regulatory element). The DRE repressed basal transcription in an orientation-independent and cell-specific manner when placed downstream from the heterologous thymidine kinase promoter. Southwestern blotting and UV cross-linking experiments with nuclear extracts from human neuroblastoma cells or human brain revealed a protein complex of approximately 110 kDa that specifically bound to the DRE. Forskolin treatment reduced binding to the DRE, and the time course paralleled that for an increase in prodynorphin gene expression. Our results suggest that under basal conditions, expression of the prodynorphin gene is repressed by occupancy of the DRE site. Upon PKA stimulation, binding to the DRE is reduced and transcription increases. We propose a model for human prodynorphin activation through PKA-dependent derepression at the DRE site.
Figures


References
-
- Adams A D, Choate D M, Thompson M A. NF1-L is the DNA-binding component of the protein complex at the peripherin negative regulatory element. J Biol Chem. 1995;270:6975–6983. - PubMed
-
- Alvarez G, Fairén A, Douglass J, Naranjo J R. Expression of the prodynorphin gene in the developing and adult cerebral cortex of the rat: an in situ hybridization study. J Comp Neurol. 1990;300:287–300. - PubMed
-
- Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: Wiley; 1995.
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
