TAL1 and LIM-only proteins synergistically induce retinaldehyde dehydrogenase 2 expression in T-cell acute lymphoblastic leukemia by acting as cofactors for GATA3
- PMID: 9819382
- PMCID: PMC109277
- DOI: 10.1128/MCB.18.12.6939
TAL1 and LIM-only proteins synergistically induce retinaldehyde dehydrogenase 2 expression in T-cell acute lymphoblastic leukemia by acting as cofactors for GATA3
Abstract
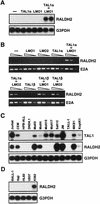
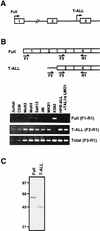
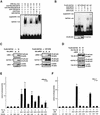
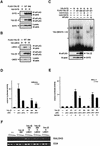
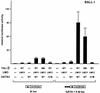
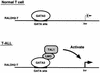
Previously, we have shown that TAL1 and the LIM-only protein gene (LMO) are regularly coactivated in T-cell acute lymphoblastic leukemia (T-ALL). This observation is likely to relate to the findings that TAL1 and LMO are highly synergistic in T-cell tumorigenesis in double-transgenic mice. To understand the molecular mechanisms of functional synergy between TAL1 and LMO in tumorigenesis and transcriptional regulation, we tried to identify downstream target genes regulated by TAL1 and LMO by a subtractive PCR method. One of the isolated genes, that for retinaldehyde dehydrogenase 2 (RALDH2), was regularly expressed in most of the T-ALL cell lines that coexpressed TAL1 and LMO. Exogenously transfected TAL1 and LMO, but not either alone, induced RALDH2 expression in a T-ALL cell line, HPB-ALL, not expressing endogeneous TAL1 or LMO. The RALDH2 transcripts in T-ALL were, however, mostly initiated within the second intron. Promoter analysis revealed that a GATA site in a cryptic promoter in the second intron was essential and sufficient for the TAL1- and LMO-dependent transcriptional activation, and GATA3 binds to this site. In addition, forced expression of GATA3 potentiated the induction of RALDH2 by TAL1 and LMO, and these three factors formed a complex in vivo. Furthermore, a TAL1 mutant not binding to DNA also activated the transcription of RALDH2 in the presence of LMO and GATA3. Collectively, we have identified the RALDH2 gene as a first example of direct transcriptional target genes regulated by TAL1 and LMO in T-ALL. In this case, TAL1 and LMO act as cofactors for GATA3 to activate the transcription of RALDH2.
Figures


Similar articles
-
Transcriptional activity of TAL1 in T cell acute lymphoblastic leukemia (T-ALL) requires RBTN1 or -2 and induces TALLA1, a highly specific tumor marker of T-ALL.J Biol Chem. 1997 Feb 14;272(7):4576-81. doi: 10.1074/jbc.272.7.4576. J Biol Chem. 1997. PMID: 9020185
-
NKX3.1 is a direct TAL1 target gene that mediates proliferation of TAL1-expressing human T cell acute lymphoblastic leukemia.J Exp Med. 2010 Sep 27;207(10):2141-56. doi: 10.1084/jem.20100745. Epub 2010 Sep 20. J Exp Med. 2010. PMID: 20855495 Free PMC article.
-
A DNA-binding mutant of TAL1 cooperates with LMO2 to cause T cell leukemia in mice.Oncogene. 2011 Mar 10;30(10):1252-60. doi: 10.1038/onc.2010.495. Epub 2010 Nov 8. Oncogene. 2011. PMID: 21057528 Free PMC article.
-
Transcription factors of the bHLH and LIM families: synergistic mediators of T cell acute leukemia?Curr Top Microbiol Immunol. 1997;220:55-65. doi: 10.1007/978-3-642-60479-9_4. Curr Top Microbiol Immunol. 1997. PMID: 9103675 Review. No abstract available.
-
LMO T-cell translocation oncogenes typify genes activated by chromosomal translocations that alter transcription and developmental processes.Genes Dev. 1998 Sep 1;12(17):2651-7. doi: 10.1101/gad.12.17.2651. Genes Dev. 1998. PMID: 9732263 Review. No abstract available.
Cited by
-
TRIB2 reinforces the oncogenic transcriptional program controlled by the TAL1 complex in T-cell acute lymphoblastic leukemia.Leukemia. 2016 Apr;30(4):959-62. doi: 10.1038/leu.2015.195. Epub 2015 Jul 23. Leukemia. 2016. PMID: 26202930 Free PMC article. No abstract available.
-
MiR-146b negatively regulates migration and delays progression of T-cell acute lymphoblastic leukemia.Sci Rep. 2016 Aug 23;6:31894. doi: 10.1038/srep31894. Sci Rep. 2016. PMID: 27550837 Free PMC article.
-
Gata2, Fli1, and Scl form a recursively wired gene-regulatory circuit during early hematopoietic development.Proc Natl Acad Sci U S A. 2007 Nov 6;104(45):17692-7. doi: 10.1073/pnas.0707045104. Epub 2007 Oct 25. Proc Natl Acad Sci U S A. 2007. PMID: 17962413 Free PMC article.
-
Retinoid metabolism and ALDH1A2 (RALDH2) expression are altered in the transgenic adenocarcinoma mouse prostate model.Biochem Pharmacol. 2009 Nov 1;78(9):1127-38. doi: 10.1016/j.bcp.2009.06.022. Epub 2009 Jun 21. Biochem Pharmacol. 2009. PMID: 19549509 Free PMC article.
-
LMO2 at 25 years: a paradigm of chromosomal translocation proteins.Open Biol. 2015 Jun;5(6):150062. doi: 10.1098/rsob.150062. Open Biol. 2015. PMID: 26108219 Free PMC article. Review.
References
-
- Agulnick A D, Taira M, Breen J J, Tanaka T, Dawid I B, Westphal H. Interactions of the LIM-domain-binding factor Ldb1 with LIM homeodomain proteins. Nature (London) 1996;384:270–272. - PubMed
-
- Bach I, Carriere C, Ostendorff H P, Andersen B, Rosenfeld M G. A family of LIM domain-associated cofactors confer transcriptional synergism between LIM and Otx homeodomain proteins. Genes Dev. 1997;11:1370–1380. - PubMed
-
- Bash R O, Hall S, Timmons C F, Crist W M, Amylon M, Smith R G, Baer R. Does activation of the TAL1 gene occur in a majority of patients with T-cell acute lymphoblastic leukemia? A Pediatric Oncology Group study. Blood. 1995;86:666–676. - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
