Regulation of RasGRP via a phorbol ester-responsive C1 domain
- PMID: 9819387
- PMCID: PMC109282
- DOI: 10.1128/MCB.18.12.6995
Regulation of RasGRP via a phorbol ester-responsive C1 domain
Abstract
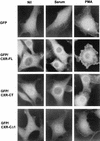
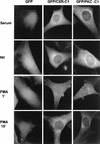
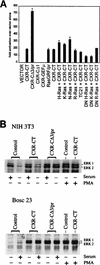
As part of a cDNA library screen for clones that induce transformation of NIH 3T3 fibroblasts, we have isolated a cDNA encoding the murine homolog of the guanine nucleotide exchange factor RasGRP. A point mutation predicted to prevent interaction with Ras abolished the ability of murine RasGRP (mRasGRP) to transform fibroblasts and to activate mitogen-activated protein kinases (MAP kinases). MAP kinase activation via mRasGRP was enhanced by coexpression of H-, K-, and N-Ras and was partially suppressed by coexpression of dominant negative forms of H- and K-Ras. The C terminus of mRasGRP contains a pair of EF hands and a C1 domain which is very similar to the phorbol ester- and diacylglycerol-binding C1 domains of protein kinase Cs. The EF hands could be deleted without affecting the ability of mRasGRP to transform NIH 3T3 cells. In contrast, deletion of the C1 domain or an adjacent cluster of basic amino acids eliminated the transforming activity of mRasGRP. Transformation and MAP kinase activation via mRasGRP were restored if the deleted C1 domain was replaced either by a membrane-localizing prenylation signal or by a diacylglycerol- and phorbol ester-binding C1 domain of protein kinase C. The transforming activity of mRasGRP could be regulated by phorbol ester when serum concentrations were low, and this effect of phorbol ester was dependent on the C1 domain of mRasGRP. The C1 domain could also confer phorbol myristate acetate-regulated transforming activity on a prenylation-defective mutant of K-Ras. The C1 domain mediated the translocation of mRasGRP to cell membranes in response to either phorbol ester or serum stimulation. These results suggest that the primary mechanism of activation of mRasGRP in fibroblasts is through its recruitment to diacylglycerol-enriched membranes. mRasGRP is expressed in lymphoid tissues and the brain, as well as in some lymphoid cell lines. In these cells, RasGRP has the potential to serve as a direct link between receptors which stimulate diacylglycerol-generating phospholipase Cs and the activation of Ras.
Figures






References
-
- Aronheim A, Engelberg D, Li N, Al-Alawi N, Schlessinger J, Karin M. Membrane targeting of the nucleotide exchange factor Sos is sufficient for activating the Ras signaling pathway. Cell. 1994;78:949–961. - PubMed
-
- Boguski M S, McCormick F. Proteins regulating Ras and its relatives. Nature. 1993;366:643–654. - PubMed
-
- Bokoch G M, Der C J. Emerging concepts in the Ras superfamily of GTP-binding proteins. FASEB J. 1993;7:750–759. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
