A functional homolog of a yeast tRNA splicing enzyme is conserved in higher eukaryotes and in Escherichia coli
- PMID: 9826666
- PMCID: PMC24339
- DOI: 10.1073/pnas.95.24.14136
A functional homolog of a yeast tRNA splicing enzyme is conserved in higher eukaryotes and in Escherichia coli
Abstract
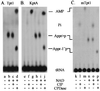
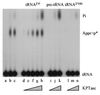
tRNA splicing in the yeast Saccharomyces cerevisiae requires an endonuclease to excise the intron, tRNA ligase to join the tRNA half-molecules, and 2'-phosphotransferase to transfer the splice junction 2'-phosphate from ligated tRNA to NAD, producing ADP ribose 1"-2" cyclic phosphate (Appr>p). We show here that functional 2'-phosphotransferases are found throughout eukaryotes, occurring in two widely divergent yeasts (Candida albicans and Schizosaccharomyces pombe), a plant (Arabidopsis thaliana), and mammals (Mus musculus); this finding is consistent with a role for the enzyme, acting in concert with ligase, to splice tRNA or other RNA molecules. Surprisingly, functional 2'-phosphotransferase is found also in the bacterium Escherichia coli, which does not have any known introns of this class, and does not appear to have a ligase that generates junctions with a 2'-phosphate. Analysis of the database shows that likely members of the 2'-phosphotransferase family are found also in one other bacterium (Pseudomonas aeruginosa) and two archaeal species (Archaeoglobus fulgidus and Pyrococcus horikoshii). Phylogenetic analysis reveals no evidence for recent horizontal transfer of the 2'-phosphotransferase into Eubacteria, suggesting that the 2'-phosphotransferase has been present there since close to the time that the three kingdoms diverged. Although 2'-phosphotransferase is not present in all Eubacteria, and a gene disruption experiment demonstrates that the protein is not essential in E. coli, the continued presence of 2'-phosphotransferase in Eubacteria over large evolutionary times argues for an important role for the protein.
Figures


Similar articles
-
Substrate recognition by a yeast 2'-phosphotransferase involved in tRNA splicing and by its Escherichia coli homolog.Biochemistry. 2001 Nov 20;40(46):14098-105. doi: 10.1021/bi011388t. Biochemistry. 2001. PMID: 11705403
-
A 2'-phosphotransferase implicated in tRNA splicing is essential in Saccharomyces cerevisiae.J Biol Chem. 1997 May 16;272(20):13203-10. doi: 10.1074/jbc.272.20.13203. J Biol Chem. 1997. PMID: 9148937
-
Analysis of 2'-phosphotransferase (Tpt1p) from Saccharomyces cerevisiae: evidence for a conserved two-step reaction mechanism.RNA. 2005 Jan;11(1):99-106. doi: 10.1261/rna.7194605. RNA. 2005. PMID: 15611300 Free PMC article.
-
RNA-splicing endonuclease structure and function.Cell Mol Life Sci. 2008 Apr;65(7-8):1176-85. doi: 10.1007/s00018-008-7393-y. Cell Mol Life Sci. 2008. PMID: 18217203 Free PMC article. Review.
-
Recent insights into the structure, function, and regulation of the eukaryotic transfer RNA splicing endonuclease complex.Wiley Interdiscip Rev RNA. 2022 Sep;13(5):e1717. doi: 10.1002/wrna.1717. Epub 2022 Feb 14. Wiley Interdiscip Rev RNA. 2022. PMID: 35156311 Free PMC article. Review.
Cited by
-
Diversity and roles of (t)RNA ligases.Cell Mol Life Sci. 2012 Aug;69(16):2657-70. doi: 10.1007/s00018-012-0944-2. Epub 2012 Mar 17. Cell Mol Life Sci. 2012. PMID: 22426497 Free PMC article. Review.
-
Chemical synthesis of 2″OMeNAD+ and its deployment as an RNA 2'-phosphotransferase (Tpt1) 'poison' that traps the enzyme on its abortive RNA-2'-PO4-(ADP-2″OMe-ribose) reaction intermediate.Nucleic Acids Res. 2024 Sep 23;52(17):10533-10542. doi: 10.1093/nar/gkae695. Nucleic Acids Res. 2024. PMID: 39162230 Free PMC article.
-
Human RNA 5'-kinase (hClp1) can function as a tRNA splicing enzyme in vivo.RNA. 2008 Sep;14(9):1737-45. doi: 10.1261/rna.1142908. Epub 2008 Jul 22. RNA. 2008. PMID: 18648070 Free PMC article.
-
The life and times of a tRNA.RNA. 2023 Jul;29(7):898-957. doi: 10.1261/rna.079620.123. Epub 2023 Apr 13. RNA. 2023. PMID: 37055150 Free PMC article. Review.
-
Transfer RNA post-transcriptional processing, turnover, and subcellular dynamics in the yeast Saccharomyces cerevisiae.Genetics. 2013 May;194(1):43-67. doi: 10.1534/genetics.112.147470. Genetics. 2013. PMID: 23633143 Free PMC article. Review.
References
-
- Xu M Q, Kathe S D, Goodrich-Blair H, Nierzwicki-Bauer S A, Shub D A. Science. 1990;250:1566–1570. - PubMed
-
- Reinhold-Hurek B, Shub D A. Nature (London) 1992;357:173–176. - PubMed
-
- Ferat J L, Michel F. Nature (London) 1993;364:358–361. - PubMed
-
- Belfort M, Weiner A. Cell. 1997;89:1003–1006. - PubMed
-
- Abelson J, Trotta C R, Li H. J Biol Chem. 1998;273:12685–12688. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

