Stochastic processes strongly influence HIV-1 evolution during suboptimal protease-inhibitor therapy
- PMID: 9826719
- PMCID: PMC24392
- DOI: 10.1073/pnas.95.24.14441
Stochastic processes strongly influence HIV-1 evolution during suboptimal protease-inhibitor therapy
Abstract
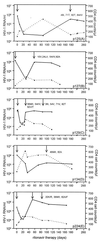
It has long been assumed that HIV-1 evolution is best described by deterministic evolutionary models because of the large population size. Recently, however, it was suggested that the effective population size (Ne) may be rather small, thereby allowing chance to influence evolution, a situation best described by a stochastic evolutionary model. To gain experimental evidence supporting one of the evolutionary models, we investigated whether the development of resistance to the protease inhibitor ritonavir affected the evolution of the env gene. Sequential serum samples from five patients treated with ritonavir were used for analysis of the protease gene and the V3 domain of the env gene. Multiple reverse transcription-PCR products were cloned, sequenced, and used to construct phylogenetic trees and to calculate the genetic variation and Ne. Genotypic resistance to ritonavir developed in all five patients, but each patient displayed a unique combination of mutations, indicating a stochastic element in the development of ritonavir resistance. Furthermore, development of resistance induced clear bottleneck effects in the env gene. The mean intrasample genetic variation, which ranged from 1.2% to 5.7% before treatment, decreased significantly (P < 0.025) during treatment. In agreement with these findings, Ne was estimated to be very small (500-15,000) compared with the total HIV-1 RNA copy number. This study combines three independent observations, strong population bottlenecking, small Ne, and selection of different combinations of protease-resistance mutations, all of which indicate that HIV-1 evolution is best described by a stochastic evolutionary model.
Figures


References
-
- Domingo E, Escarmis C, Sevilla N, Moya A, Elena S F, Quer J, Novella I S, Holland J J. FASEB J. 1996;10:859–864. - PubMed
-
- Coffin J M. Curr Top Microbiol Immunol. 1992;176:143–164. - PubMed
-
- Domingo E, Holland J J. Annu Rev Microbiol. 1997;51:151–178. - PubMed
-
- Eigen M. Steps Toward Life. Oxford: Oxford Univ. Press; 1992.
-
- Eigen M. Gene. 1993;135:37–47. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

