Role of SarA in virulence determinant production and environmental signal transduction in Staphylococcus aureus
- PMID: 9829932
- PMCID: PMC107708
- DOI: 10.1128/JB.180.23.6232-6241.1998
Role of SarA in virulence determinant production and environmental signal transduction in Staphylococcus aureus
Abstract
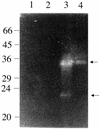
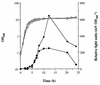
The staphylococcal accessory regulator (encoded by sarA) is an important global regulator of virulence factor biosynthesis in Staphylococcus aureus. To further characterize its role in virulence determinant production, an sarA knockout mutant was created by insertion of a kanamycin antibiotic resistance cassette into the sarA gene. N-terminal sequencing of exoproteins down-regulated by sarA identified several putative proteases, including a V8 serine protease and a novel metalloprotease, as the major extracellular proteins repressed by sarA. In kinetic studies, the sarA mutation delays the onset of alpha-hemolysin (encoded by hla) expression and reduces levels of hla to approximately 40% of the parent strain level. Furthermore, SarA plays a role in signal transduction in response to microaerobic growth since levels of hla were much lower in a microaerobic environment than after aerobic growth in the sarA mutant. An exoprotein exhibiting hemolysin activity on sheep blood, and up-regulated by sarA independently of the accessory gene regulator (encoded by agr), was specifically induced microaerobically. Transcriptional gene fusion and Western analysis revealed that sarA up-regulates both toxic shock syndrome toxin 1 gene (tst) expression and staphylococcal enterotoxin B production, respectively. This study demonstrates the role of sarA as a signal transduction regulatory component in response to aeration stimuli and suggests that sarA functions as a major repressor of protease activity. The possible role of proteases as regulators of virulence determinant stability is discussed.
Figures
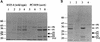



References
-
- Arvidson S. Studies on extracellular proteolytic enzymes from Staphylococcus aureus. II. Isolation and characterisation of an EDTA-sensitive protease. Biochim Biophys Acta. 1973;302:149–157. - PubMed
-
- Arvidson S O. Extracellular enzymes from Staphylococcus aureus. In: Easmon C S F, Adlam C, editors. Staphylococci and staphylococcal infections. Vol. 2. London, England: Academic Press Inc.; 1983. pp. 745–808.
-
- Ayora S, Götz F. Genetic and biochemical properties of an extracellular neutral metalloprotease from Staphylococcus hyicus subsp. hyicus. Mol Gen Genet. 1994;242:421–430. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

