Role and regulation of Bacillus subtilis glutamate dehydrogenase genes
- PMID: 9829940
- PMCID: PMC107716
- DOI: 10.1128/JB.180.23.6298-6305.1998
Role and regulation of Bacillus subtilis glutamate dehydrogenase genes
Abstract
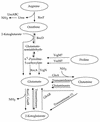
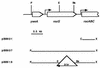
The complete Bacillus subtilis genome contains two genes with the potential to encode glutamate dehydrogenase (GlutDH) enzymes. Mutations in these genes were constructed and characterized. The rocG gene proved to encode a major GlutDH whose synthesis was induced in media containing arginine or ornithine or, to a lesser degree, proline and was repressed by glucose. A rocG null mutant was impaired in utilization of arginine, ornithine, and proline as nitrogen or carbon sources. The gudB gene was expressed under all growth conditions tested but codes for a GlutDH that seemed to be intrinsically inactive. Spontaneous mutations in gudB that removed a 9-bp direct repeat within the wild-type gudB sequence activated the GudB protein and allowed more-efficient utilization of amino acids of the glutamate family.
Figures



References
-
- Baker P J, Britton K L, Engel P C, Farrants G W, Lilley K S, Rice D W, Stillman T J. Subunit assembly and active site location in the structure of glutamate dehydrogenase. Proteins Struct Funct Genet. 1992;12:75–86. - PubMed
-
- Belitsky, B. R. Unpublished data.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

