Combined physical and genetic map of the Pseudomonas putida KT2440 chromosome
- PMID: 9829947
- PMCID: PMC107723
- DOI: 10.1128/JB.180.23.6352-6363.1998
Combined physical and genetic map of the Pseudomonas putida KT2440 chromosome
Abstract
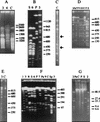
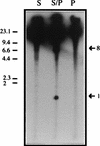
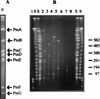
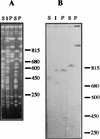
A combined physical and genetic map of the Pseudomonas putida KT2440 genome was constructed from data obtained by pulsed-field gel electrophoresis techniques (PFGE) and Southern hybridization. Circular genome size was estimated at 6.0 Mb by adding the sizes of 19 SwaI, 9 PmeI, 6 PacI, and 6 I-CeuI fragments. A complete physical map was achieved by combining the results of (i) analysis of PFGE of the DNA fragments resulting from digestion of the whole genome with PmeI, SwaI, I-CeuI, and PacI as well as double digestion with combinations of these enzymes and (ii) Southern hybridization analysis of the whole wild-type genome digested with different enzymes and hybridized against a series of probes obtained as cloned genes from different pseudomonads of rRNA group I and Escherichia coli, as P. putida DNA obtained by PCR amplification based on sequences deposited at the GenBank database, and by labeling of macrorestriction fragments of the P. putida genome eluted from agarose gels. As an alternative, 10 random mini-Tn5-Km mutants of P. putida KT2440 were used as a source of DNA, and the band carrying the mini-Tn5 in each mutant was identified after PFGE of a series of complete chromosomal digestions and hybridization with the kanamycin resistance gene of the mini-Tn5 as a probe. We established a circular genome map with an average resolution of 160 kb. Among the 63 genes located on the genetic map were key markers such as oriC, 6 rrn loci (rnnA to -F), recA, ftsZ, rpoS, rpoD, rpoN, and gyrB; auxotrophic markers; and catabolic genes for the metabolism of aromatic compounds. The genetic map of P. putida KT2440 was compared to those of Pseudomonas aeruginosa PAO1 and Pseudomonas fluorescens SBW25. The chromosomal backbone revealed some similarity in gene clustering among the three pseudomonads but differences in physical organization, probably as a result of intraspecific rearrangements.
Figures



Similar articles
-
Physical and genetic map of the Pseudomonas fluorescens SBW25 chromosome.Mol Microbiol. 1996 Feb;19(3):521-33. doi: 10.1046/j.1365-2958.1996.391926.x. Mol Microbiol. 1996. PMID: 8830243
-
Physical map of the genome of Rhizobium meliloti 1021.J Bacteriol. 1993 Nov;175(21):6945-52. doi: 10.1128/jb.175.21.6945-6952.1993. J Bacteriol. 1993. PMID: 8226638 Free PMC article.
-
A physical and genetic map of the Corynebacterium glutamicum ATCC 13032 chromosome.Mol Gen Genet. 1996 Sep 13;252(3):255-65. doi: 10.1007/BF02173771. Mol Gen Genet. 1996. PMID: 8842145
-
A PacI/SwaI map of the Pseudomonas aeruginosa PAO chromosome.Electrophoresis. 1992 Sep-Oct;13(9-10):649-51. doi: 10.1002/elps.11501301135. Electrophoresis. 1992. PMID: 1459083
-
Unraveling the genomic diversity of the Pseudomonas putida group: exploring taxonomy, core pangenome, and antibiotic resistance mechanisms.FEMS Microbiol Rev. 2024 Nov 23;48(6):fuae025. doi: 10.1093/femsre/fuae025. FEMS Microbiol Rev. 2024. PMID: 39390673 Free PMC article. Review.
Cited by
-
Presence of Pseudomonas putida strains harboring plasmids bearing the metallo-beta-lactamase gene bla(IMP) in a hospital in Japan.J Clin Microbiol. 2003 Sep;41(9):4246-51. doi: 10.1128/JCM.41.9.4246-4251.2003. J Clin Microbiol. 2003. PMID: 12958252 Free PMC article.
-
Inactivation of gltB abolishes expression of the assimilatory nitrate reductase gene (nasB) in Pseudomonas putida KT2442.J Bacteriol. 2000 Jun;182(12):3368-76. doi: 10.1128/JB.182.12.3368-3376.2000. J Bacteriol. 2000. PMID: 10852866 Free PMC article.
-
Colorimetric method for identifying plant essential oil components that affect biofilm formation and structure.Appl Environ Microbiol. 2004 Dec;70(12):6951-6. doi: 10.1128/AEM.70.12.6951-6956.2004. Appl Environ Microbiol. 2004. PMID: 15574886 Free PMC article.
-
Growth independent rhamnolipid production from glucose using the non-pathogenic Pseudomonas putida KT2440.Microb Cell Fact. 2011 Oct 17;10:80. doi: 10.1186/1475-2859-10-80. Microb Cell Fact. 2011. PMID: 21999513 Free PMC article.
-
Variable copy number, intra-genomic heterogeneities and lateral transfers of the 16S rRNA gene in Pseudomonas.PLoS One. 2012;7(4):e35647. doi: 10.1371/journal.pone.0035647. Epub 2012 Apr 24. PLoS One. 2012. PMID: 22545126 Free PMC article.
References
-
- Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1991.
-
- Bautsch W, Grothues D, Tümmler B. Genome fingerprinting of Pseudomonas aeruginosa by two dimensional field inversion gel electrophoresis. FEMS Microbiol Lett. 1988;52:255–258.
-
- Calero S, Garriga X, Barbé J. Analysis of the DNA damage-mediated induction of Pseudomonas putida and Pseudomonas aeruginosa lexA gene. FEMS Microbiol Lett. 1993;110:65–70. - PubMed
-
- Challis B C. Molecular characterization of the C4-dicarboxylate transport system of P. fluorescens and its role in root colonization. M.Sc. thesis. Dunedin, New Zealand: University of Otago; 1994.
-
- Cole S T, Saint Girons I. Bacterial genomics. FEMS Microbiol Rev. 1994;14:139–160. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

