Identification and characterization of the Myxococcus xanthus argE gene
- PMID: 9829957
- PMCID: PMC107734
- DOI: 10.1128/JB.180.23.6412-6414.1998
Identification and characterization of the Myxococcus xanthus argE gene
Abstract
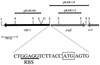
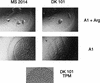
The chromosomal acetylornithine deacetylase (argE) gene of Myxococcus xanthus was identified via homology to acetylornithine deacetylases from other bacterial species. A mutant carrying a disruption in argE was unable to grow on minimal media lacking supplemental arginine and formed fruiting bodies and spores in response to arginine starvation at high cell density.
Figures


References
-
- Bibb M J, Findlay P R, Johnson M W. The relationship between base composition and codon usage in bacterial genes and its use for the simple and reliable identification of protein coding sequences. Gene. 1984;30:157–166. - PubMed
-
- Glansdorff N. Biosynthesis of arginine and polyamines. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 408–433.
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

