The Oct-1 POU domain activates snRNA gene transcription by contacting a region in the SNAPc largest subunit that bears sequence similarities to the Oct-1 coactivator OBF-1
- PMID: 9832505
- PMCID: PMC317248
- DOI: 10.1101/gad.12.22.3528
The Oct-1 POU domain activates snRNA gene transcription by contacting a region in the SNAPc largest subunit that bears sequence similarities to the Oct-1 coactivator OBF-1
Abstract
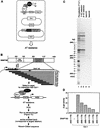
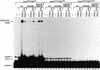
The RNA polymerases II and III snRNA gene promoters contain an octamer sequence as part of the enhancer and a proximal sequence element (PSE) as part of the core promoter. The octamer and the PSE bind the POU domain activator Oct-1 and the basal transcription factor SNAPc, respectively. Oct-1, but not Oct-1 with a single E7R mutation within the POU domain, binds cooperatively with SNAPc and, in effect, recruits SNAPc to the PSE. Here, we show that SNAPc recruitment is mediated by an interaction between the Oct-1 POU domain and a small region of the largest subunit of SNAPc, SNAP190. This SNAP190 region is strikingly similar to a region in the B-cell-specific Oct-1 coactivator, OBF-1, that is required for interaction with octamer-bound Oct-1 POU domain. The Oct-1 POU domain-SNAP190 interaction is a direct protein-protein contact as determined by the isolation of a switched specificity SNAP190 mutant that interacts with Oct-1 POU E7R but not with wild-type Oct-1 POU. We also show that this direct protein-protein contact results in activation of transcription. Thus, we have identified an activation target of a human activator, Oct-1, within its cognate basal transcription complex.
Figures

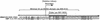


References
-
- Assa-Munt N, Mortishire-Smith RJ, Aurora R, Herr W, Wright PE. The solution structure of the Oct-1 POU-specific domain reveals a striking similarity to the bacteriophage lamda repressor DNA-binding domain. Cell. 1993;73:193–205. - PubMed
-
- Beamer LJ, Pabo CO. Refined 1.8 A crystal structure of the lambda repressor-operator complex. J Mol Biol. 1992;227:177–196. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
