The molecular mechanism of mitotic inhibition of TFIIH is mediated by phosphorylation of CDK7
- PMID: 9832506
- PMCID: PMC317239
- DOI: 10.1101/gad.12.22.3541
The molecular mechanism of mitotic inhibition of TFIIH is mediated by phosphorylation of CDK7
Abstract
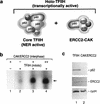
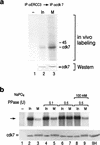
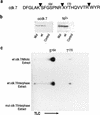
TFIIH is a multisubunit complex, containing ATPase, helicases, and kinase activities. Functionally, TFIIH has been implicated in transcription by RNA polymerase II (RNAPII) and in nucleotide excision repair. A member of the cyclin-dependent kinase family, CDK7, is the kinase subunit of TFIIH. Genetically, CDK7 homologues have been implicated in transcription in Saccharomyces cerevisiae, and in mitotic regulation in Schizosaccharomyces pombe. Here we show that in mitosis the CDK7 subunit of TFIIH and the largest subunit of RNAPII become hyperphosphorylated. MPF-induced phosphorylation of CDK7 results in inhibition of the TFIIH-associated kinase and transcription activities. Negative and positive regulation of TFIIH requires phosphorylation within the T-loop of CDK7. Our data establishes TFIIH and its subunit CDK7 as a direct link between the regulation of transcription and the cell cycle.
Figures




References
-
- Akoulitchev S, Mäkelä TP, Weinberg RA, Reinberg D. Requirement for TFIIH kinase activity in transcription by RNA polymerase II. Nature. 1995;377:557–560. - PubMed
-
- Cisek LJ, Corden JL. Phosphorylation of RNA polymerase by the murine homologue of the cell-cycle control protein cdc2. Nature. 1989;339:679–684. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
