Dorsoventral patterning in the Drosophila central nervous system: the intermediate neuroblasts defective homeobox gene specifies intermediate column identity
- PMID: 9832510
- PMCID: PMC317240
- DOI: 10.1101/gad.12.22.3591
Dorsoventral patterning in the Drosophila central nervous system: the intermediate neuroblasts defective homeobox gene specifies intermediate column identity
Abstract
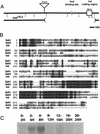
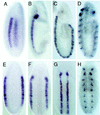
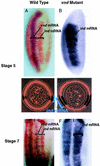
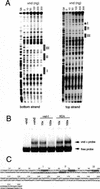
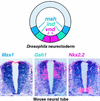
One of the first steps in neurogenesis is the diversification of cells along the dorsoventral axis. In Drosophila the central nervous system develops from three longitudinal columns of cells: ventral cells that express the vnd/nk2 homeobox gene, intermediate cells, and dorsal cells that express the msh homeobox gene. Here we describe a new Drosophila homeobox gene, intermediate neuroblasts defective (ind), which is expressed specifically in the intermediate column cells. ind is essential for intermediate column development: Null mutants have a transformation of intermediate to dorsal column neuroectoderm fate, and only 10% of the intermediate column neuroblasts develop. The establishment of dorsoventral column identity involves negative regulation: Vnd represses ind in the ventral column, whereas ind represses msh in the intermediate column. Vertebrate genes closely related to vnd (Nkx2.1 and Nkx2.2), ind (Gsh1 and Gsh2), and msh (Msx1 and Msx3) are expressed in corresponding ventral, intermediate, and dorsal domains during vertebrate neurogenesis, raising the possibility that dorsoventral patterning within the central nervous system is evolutionarily conserved.
Figures


References
-
- Adams B, Dorfler P, Aguzzi A, Kozmik Z, Urbanek P, Maurer-Fogy I, Busslinger M. Pax-5 encodes the transcription factor BSAP and is expressed in B lymphocytes, the developing CNS, and adult testis. Genes & Dev. 1992;6:1589–1607. - PubMed
-
- Angerer L, Angerer R. In situ hybridization to cellular RNA with radiolabelled RNA probes. In: Wilkinson DG, editor. In situ hybridization: A practical approach. Oxford, UK: Oxford University Press; 1992. pp. 15–32.
-
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current protocols in molecular biology. New York, NY: John Wiley & Sons; 1994.
-
- Azpiazu N, Frasch M. tinman and bagpipe: Two homeobox genes that determine cell fates in the dorsal mesoderm of Drosophila. Genes & Dev. 1993;7:1325–1340. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
