An Escherichia coli expression assay and screen for human immunodeficiency virus protease variants with decreased susceptibility to indinavir
- PMID: 9835523
- PMCID: PMC106031
- DOI: 10.1128/AAC.42.12.3256
An Escherichia coli expression assay and screen for human immunodeficiency virus protease variants with decreased susceptibility to indinavir
Abstract
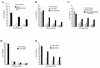
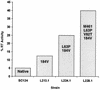
We have developed a recombinant Escherichia coli screening system for the rapid detection and identification of amino acid substitutions in the human immunodeficiency virus (HIV) protease associated with decreased susceptibility to the protease inhibitor indinavir (MK-639; Merck & Co.). The assay depends upon the correct processing of a segment of the HIV-1 HXB2 gag-pol polyprotein followed by detection of HIV reverse transcriptase activity by a highly sensitive, colorimetric enzyme-linked immunosorbent assay. The highly sensitive system detects the contributions of single substitutions such as I84V, L90M, and L63P. The combination of single substitutions further decreases the sensitivity to indinavir. We constructed a library of HIV protease variant genes containing dispersed mutations and, using the E. coli recombinant system, screened for mutants with decreased indinavir sensitivity. The discovered HIV protease variants contain amino acid substitutions commonly associated with indinavir resistance in clinical isolates, including the substitutions L90M, L63P, I64V, V82A, L24I, and I54T. One substitution, W6R, is also frequently found by the screen and has not been reported elsewhere. Of a total of 12,000 isolates that were screened, 12 protease variants with decreased sensitivity to indinavir were found. The L63P substitution, which is also associated with indinavir resistance, increases the stability of the isolated protease relative to that of the native HXB2 protease. The rapidity, sensitivity, and accuracy of this screen also make it useful for screening for novel inhibitors. We have found the approach described here to be useful for the detection of amino acid substitutions in HIV protease that have been associated with drug resistance as well as for the screening of novel compounds for inhibitory activity.
Figures


Similar articles
-
Crystal structures of HIV protease V82A and L90M mutants reveal changes in the indinavir-binding site.Eur J Biochem. 2004 Apr;271(8):1516-24. doi: 10.1111/j.1432-1033.2004.04060.x. Eur J Biochem. 2004. PMID: 15066177
-
Rapid phenotypic assay for human immunodeficiency virus type 1 protease using in vitro translation.J Virol Methods. 2002 Oct;106(1):25-37. doi: 10.1016/s0166-0934(02)00133-7. J Virol Methods. 2002. PMID: 12367727
-
Mutational anatomy of an HIV-1 protease variant conferring cross-resistance to protease inhibitors in clinical trials. Compensatory modulations of binding and activity.J Biol Chem. 1996 Dec 13;271(50):31957-63. doi: 10.1074/jbc.271.50.31957. J Biol Chem. 1996. PMID: 8943242
-
The influence of protease inhibitor resistance profiles on selection of HIV therapy in treatment-naive patients.Antivir Ther. 2004 Jun;9(3):301-14. Antivir Ther. 2004. PMID: 15259893 Review.
-
In vivo selection of HIV-1 variants with reduced susceptibility to the protease inhibitor L-735,524 and related compounds.Adv Exp Med Biol. 1996;394:327-31. doi: 10.1007/978-1-4757-9209-6_30. Adv Exp Med Biol. 1996. PMID: 8815697 Review.
Cited by
-
Natural polymorphisms and unusual mutations in HIV-1 protease with potential antiretroviral resistance: a bioinformatic analysis.BMC Bioinformatics. 2014 Mar 15;15:72. doi: 10.1186/1471-2105-15-72. BMC Bioinformatics. 2014. PMID: 24629078 Free PMC article.
-
Development of a novel screen for protease inhibitors.Clin Diagn Lab Immunol. 2001 Mar;8(2):437-40. doi: 10.1128/CDLI.8.2.437-440.2001. Clin Diagn Lab Immunol. 2001. PMID: 11238235 Free PMC article.
-
Discovery of amyloid-beta aggregation inhibitors using an engineered assay for intracellular protein folding and solubility.Protein Sci. 2009 Feb;18(2):277-86. doi: 10.1002/pro.33. Protein Sci. 2009. PMID: 19177561 Free PMC article.
References
-
- Cadwell R C, Joyce G F. Mutagenic PCR. PCR Methods Appl. 1994;3:S136–S140. - PubMed
-
- Cadwell R C, Joyce G F. Randomization of genes by PCR mutagenesis. PCR Methods Appl. 1992;2:28–33. - PubMed
-
- Cheng Y S, McGowan M H, Kettner C A, Schloss J V, Erickson-Viitanen S, Yin F H. High-level synthesis of recombinant HIV-1 protease and the recovery of active enzyme from inclusion bodies. Gene. 1990;87:243–248. - PubMed
-
- Condra J H, Holder D J, Schleif W A, Blahy O M, Danovich R M, Gabryelski L J, Graham D J, Laird D, Quintero J C, Rhodes A, Robbins H L, Roth E, Shivaprakash M, Yang T, Chodakewitz J A, Deutsch P J, Leavitt R Y, Massari F E, Mellors J W, Squires K E, Steigbigel R T, Teppler H, Emini E A. Genetic correlates of in vivo viral resistance to indinavir, a human immunodeficiency virus type 1 protease inhibitor. J Virol. 1996;70:8270–8276. - PMC - PubMed
-
- Condra J H, Schleif W A, Blahy O M, Gabryelski L J, Graham D J, Quintero J C, Rhodes A, Robbins H L, Roth E, Shivaprakash M, Titus D, Yang T, Teppler H, Squires K E, Deutsch P J, Emini E A. In vivo emergence of HIV-1 variants resistant to multiple protease inhibitors. Nature. 1995;374:569–571. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

