A polyphasic approach To study the diversity and vertical distribution of sulfur-oxidizing thiomicrospira species in coastal sediments of the german wadden Sea
- PMID: 9835544
- PMCID: PMC90904
- DOI: 10.1128/AEM.64.12.4650-4657.1998
A polyphasic approach To study the diversity and vertical distribution of sulfur-oxidizing thiomicrospira species in coastal sediments of the german wadden Sea
Abstract
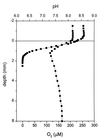
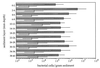
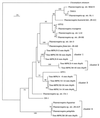
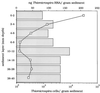
Recently, four Thiomicrospira strains were isolated from a coastal mud flat of the German Wadden Sea (T. Brinkhoff and G. Muyzer, Appl. Environ. Microbiol. 63:3789-3796, 1997). Here we describe the use of a polyphasic approach to investigate the functional role of these closely related bacteria. Microsensor measurements showed that there was oxygen penetration into the sediment to a depth of about 2.0 mm. The pH decreased from 8.15 in the overlaying water to a minimum value of 7.3 at a depth of 1.2 mm. Further down in the sediment the pH increased to about 7.8 and remained constant. Most-probable-number (MPN) counts of chemolithoautotrophic sulfur-oxidizing bacteria revealed nearly constant numbers along the vertical profile; the cell concentration ranged from 0.93 x 10(5) to 9.3 x 10(5) cells per g of sediment. A specific PCR was used to detect the presence of Thiomicrospira cells in the MPN count preparations and to determine their 16S rRNA sequences. The concentration of Thiomicrospira cells did not decrease with depth. It was found that Thiomicrospira strains were not dominant sulfur-oxidizing bacteria in this habitat. Denaturing gradient gel electrophoresis (DGGE) of PCR-amplified 16S ribosomal DNA fragments followed by hybridization analysis with a genus-specific oligonucleotide probe revealed the diversity of Thiomicrospira strains in the MPN cultures. Sequence analysis of the highest MPN dilutions in which the genus Thiomicrospira was detected revealed that there were four clusters of several closely related sequences. Only one of the 10 Thiomicrospira sequences retrieved was related to sequences of known isolates from the same habitat. Slot blot hybridization of rRNA isolated from different sediment layers showed that, in contrast to the concentration of Thiomicrospira cells, the concentration of Thiomicrospira-specific rRNA decreased rapidly in the region below the oxic layer of the sediment. This study revealed the enormous sequence diversity of closely related microorganisms present in one habitat, which so far has been found only by sequencing molecular isolates. In addition, it showed that most of the Thiomicrospira populations in the sediment studied were quiescent.
Figures

References
-
- Amann R I, Kühl M. In situ methods for assessment of microorganisms and their activities. Curr Opin Microbiol. 1998;1:352–358. - PubMed
-
- American Public Health Association. Standard methods for the examination of water and wastewater, including bottom sediments and sludge. Washington, D.C: American Public Health Association; 1969. pp. 604–609.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

