A new approach to utilize PCR-single-strand-conformation polymorphism for 16S rRNA gene-based microbial community analysis
- PMID: 9835576
- PMCID: PMC90936
- DOI: 10.1128/AEM.64.12.4870-4876.1998
A new approach to utilize PCR-single-strand-conformation polymorphism for 16S rRNA gene-based microbial community analysis
Abstract
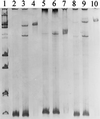
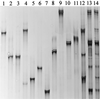
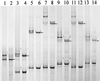
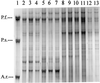
Single-strand-conformation polymorphism (SSCP) of DNA, a method widely used in mutation analysis, was adapted to the analysis and differentiation of cultivated pure-culture soil microorganisms and noncultivated rhizosphere microbial communities. A fragment (approximately 400 bp) of the bacterial 16S rRNA gene (V-4 and V-5 regions) was amplified by PCR with universal primers, with one primer phosphorylated at the 5' end. The phosphorylated strands of the PCR products were selectively digested with lambda exonuclease, and the remaining strands were separated by electrophoresis with an MDE polyacrylamide gel, a matrix specifically optimized for SSCP purposes. By this means, reannealing and heteroduplex formation of DNA strands during electrophoresis could be excluded, and the number of bands per organism was reduced. PCR products from 10 of 11 different bacterial type strains tested could be differentiated from each other. With template mixtures consisting of pure-culture DNAs from 5 and 10 bacterial strains, most of the single strains could be detected from such model communities after PCR and SSCP analyses. Purified bands amplified from pure cultures and model communities extracted from gels could be reamplified by PCR, but by this process, additional products were also generated, as detected by further SSCP analysis. Profiles generated with DNAs of rhizosphere bacterial communities, directly extracted from two different plant species grown in the same field site, could be clearly distinguished. This study demonstrates the potential of the selected PCR-single-stranded DNA approach for microbial community analysis.
Figures
References
-
- Bassam B J, Caetano-Anolles G, Gresshoff P M. Fast and sensitive silver staining of DNA in polyacrylamide gels. Anal Biochem. 1991;80:81–84. - PubMed
-
- Clayton R A, Sutton G, Hinkle P S, Jr, Bult C, Fields C. Intraspecific variation in small-subunit rRNA sequences in GenBank: why single sequences may not adequately represent prokaryotic taxa. Int J Syst Bacteriol. 1995;45:595–599. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

