Analysis of the dynamics of bacterial communities in the rhizosphere of the chrysanthemum via denaturing gradient gel electrophoresis and substrate utilization patterns
- PMID: 9835588
- PMCID: PMC90948
- DOI: 10.1128/AEM.64.12.4950-4957.1998
Analysis of the dynamics of bacterial communities in the rhizosphere of the chrysanthemum via denaturing gradient gel electrophoresis and substrate utilization patterns
Abstract
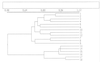
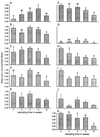
In order to gain a better understanding of the spatial and temporal dynamics of bacterial communities of the rhizosphere of the chrysanthemum, two complementary methods were used: a molecular bacterial community profiling method, i.e., 16S rRNA gene-based PCR followed by denaturing gradient gel electrophoresis (DGGE), and an agar plate method in which 11 sole-carbon-source utilization tests were used. The DGGE patterns showed that the bacterial communities as determined from direct rhizosphere DNA extracts were largely stable along developing roots of the chrysanthemum, with very little change over time or between root parts of different ages. The patterns were also similar to those produced with DNA extracts obtained from bulk soil samples. The DGGE patterns obtained by using microbial colonies from dilution plates as the source of target DNA were different from those found with the direct DNA extracts. Moreover, these patterns showed differences among plant replicates but also among replicate plates. Results obtained with the sole-carbon-source utilization tests indicated that the metabolic profile of the bacterial communities in the rhizosphere of the root tip did not change substantially during plant growth. This suggests selective development of specific bacterial populations by the presence of a root tip. On the other hand, the metabolic profile of bacterial communities in the rhizosphere of the root base changed during plant growth. With eight sole-carbon-source utilization tests, a significant effect of the development stage of the plant on the number of bacteria which were able to grow on these carbon sources was observed.
Figures





References
-
- Bowen G D, Rovira A D. The rhizosphere, the hidden half. In: Waisel Y, Eshel A, Kafkafi U, editors. Plant roots—the hidden half. New York, N.Y: Marcel Dekker; 1991. pp. 641–649.
-
- Curl E A, Truelove B. The rhizosphere. New York, N.Y: Springer; 1986.
-
- De Leij F A A M, Whipps J M, Lynch J M. The use of colony development for the characterization of bacterial communities in soil and roots. Microb Ecol. 1993;27:81–97. - PubMed
-
- Felske A, Rheims H, Wolterink A, Stackebrandt E, Akkermans A D L. Ribosome analysis reveals prominent activity of an uncultured member of the class Actinobacteria in grassland soils. Microbiology. 1997;143:2983–2989. - PubMed
LinkOut - more resources
Full Text Sources

