Molecular biology and pathogenicity of mycoplasmas
- PMID: 9841667
- PMCID: PMC98941
- DOI: 10.1128/MMBR.62.4.1094-1156.1998
Molecular biology and pathogenicity of mycoplasmas
Abstract
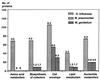
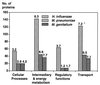
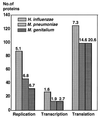
The recent sequencing of the entire genomes of Mycoplasma genitalium and M. pneumoniae has attracted considerable attention to the molecular biology of mycoplasmas, the smallest self-replicating organisms. It appears that we are now much closer to the goal of defining, in molecular terms, the entire machinery of a self-replicating cell. Comparative genomics based on comparison of the genomic makeup of mycoplasmal genomes with those of other bacteria, has opened new ways of looking at the evolutionary history of the mycoplasmas. There is now solid genetic support for the hypothesis that mycoplasmas have evolved as a branch of gram-positive bacteria by a process of reductive evolution. During this process, the mycoplasmas lost considerable portions of their ancestors' chromosomes but retained the genes essential for life. Thus, the mycoplasmal genomes carry a high percentage of conserved genes, greatly facilitating gene annotation. The significant genome compaction that occurred in mycoplasmas was made possible by adopting a parasitic mode of life. The supply of nutrients from their hosts apparently enabled mycoplasmas to lose, during evolution, the genes for many assimilative processes. During their evolution and adaptation to a parasitic mode of life, the mycoplasmas have developed various genetic systems providing a highly plastic set of variable surface proteins to evade the host immune system. The uniqueness of the mycoplasmal systems is manifested by the presence of highly mutable modules combined with an ability to expand the antigenic repertoire by generating structural alternatives, all compressed into limited genomic sequences. In the absence of a cell wall and a periplasmic space, the majority of surface variable antigens in mycoplasmas are lipoproteins. Apart from providing specific antimycoplasmal defense, the host immune system is also involved in the development of pathogenic lesions and exacerbation of mycoplasma induced diseases. Mycoplasmas are able to stimulate as well as suppress lymphocytes in a nonspecific, polyclonal manner, both in vitro and in vivo. As well as to affecting various subsets of lymphocytes, mycoplasmas and mycoplasma-derived cell components modulate the activities of monocytes/macrophages and NK cells and trigger the production of a wide variety of up-regulating and down-regulating cytokines and chemokines. Mycoplasma-mediated secretion of proinflammatory cytokines, such as tumor necrosis factor alpha, interleukin-1 (IL-1), and IL-6, by macrophages and of up-regulating cytokines by mitogenically stimulated lymphocytes plays a major role in mycoplasma-induced immune system modulation and inflammatory responses.
Figures
References
-
- Aguila H N, Lai W C, Lu Y S, Pakes S P. Experimental Mycoplasma pulmonis infection of rats suppresses humoral but not cellular immune response. Lab Anim Sci. 1988;38:138–142. - PubMed
-
- Al-Daccak R, Mehindate K, Poubelle P E, Mourad W. Signalling via MHC class II molecules selectively induces IL-1 beta over IL-1 receptor antagonist gene expression. Biochem Biophys Res Commun. 1994;201:855–860. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

