Life below the gum line: pathogenic mechanisms of Porphyromonas gingivalis
- PMID: 9841671
- PMCID: PMC98945
- DOI: 10.1128/MMBR.62.4.1244-1263.1998
Life below the gum line: pathogenic mechanisms of Porphyromonas gingivalis
Abstract
Porphyromonas gingivalis, a gram-negative anaerobe, is a major etiological agent in the initiation and progression of severe forms of periodontal disease. An opportunistic pathogen, P. gingivalis can also exist in commensal harmony with the host, with disease episodes ensuing from a shift in the ecological balance within the complex periodontal microenvironment. Colonization of the subgingival region is facilitated by the ability to adhere to available substrates such as adsorbed salivary molecules, matrix proteins, epithelial cells, and bacteria that are already established as a biofilm on tooth and epithelial surfaces. Binding to all of these substrates may be mediated by various regions of P. gingivalis fimbrillin, the structural subunit of the major fimbriae. P. gingivalis is an asaccharolytic organism, with a requirement for hemin (as a source of iron) and peptides for growth. At least three hemagglutinins and five proteinases are produced to satisfy these requirements. The hemagglutinin and proteinase genes contain extensive regions of highly conserved sequences, with posttranslational processing of proteinase gene products contributing to the formation of multimeric surface protein-adhesin complexes. Many of the virulence properties of P. gingivalis appear to be consequent to its adaptations to obtain hemin and peptides. Thus, hemagglutinins participate in adherence interactions with host cells, while proteinases contribute to inactivation of the effector molecules of the immune response and to tissue destruction. In addition to direct assault on the periodontal tissues, P. gingivalis can modulate eucaryotic cell signal transduction pathways, directing its uptake by gingival epithelial cells. Within this privileged site, P. gingivalis can replicate and impinge upon components of the innate host defense. Although a variety of surface molecules stimulate production of cytokines and other participants in the immune response, P. gingivalis may also undertake a stealth role whereby pivotal immune mediators are selectively inactivated. In keeping with its strict metabolic requirements, regulation of gene expression in P. gingivalis can be controlled at the transcriptional level. Finally, although periodontal disease is localized to the tissues surrounding the tooth, evidence is accumulating that infection with P. gingivalis may predispose to more serious systemic conditions such as cardiovascular disease and to delivery of preterm infants.
Figures


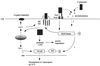
 ; pathway with potential
intermediate steps;
; pathway with potential
intermediate steps;  ,
translocation;
,
translocation;  , release;
, release;
 , reversible association.
Compiled from references , , , , , ,
, and .
, reversible association.
Compiled from references , , , , , ,
, and .
References
-
- Allaker R P, Aduse-Opoku J, Batten J E, Curtis M A. Natural variation within the principal arginine-specific protease gene, prpR1, of Porphyromonas gingivalis. Oral Microbiol Immunol. 1997;12:298–302. - PubMed
-
- Amano A, Fujiwara T, Nagata H, Kuboniwa M, Shamra A, Sojar H T, Genco R J, Hamada S, Shizukuishi S. Porphyromonas gingivalis fimbriae mediate coaggregation with Streptococcus oralisthrough specific domains. J Dent Res. 1997;76:852–857. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

