A natural view of microbial biodiversity within hot spring cyanobacterial mat communities
- PMID: 9841675
- PMCID: PMC98949
- DOI: 10.1128/MMBR.62.4.1353-1370.1998
A natural view of microbial biodiversity within hot spring cyanobacterial mat communities
Abstract
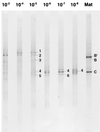
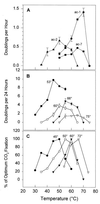
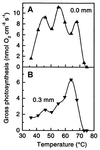
This review summarizes a decade of research in which we have used molecular methods, in conjunction with more traditional approaches, to study hot spring cyanobacterial mats as models for understanding principles of microbial community ecology. Molecular methods reveal that the composition of these communities is grossly oversimplified by microscopic and cultivation methods. For example, none of 31 unique 16S rRNA sequences detected in the Octopus Spring mat, Yellowstone National Park, matches that of any prokaryote previously cultivated from geothermal systems; 11 are contributed by genetically diverse cyanobacteria, even though a single cyanobacterial species was suspected based on morphologic and culture analysis. By studying the basis for the incongruity between culture and molecular samplings of community composition, we are beginning to cultivate isolates whose 16S rRNA sequences are readily detected. By placing the genetic diversity detected in context with the well-defined natural environmental gradients typical of hot spring mat systems, the relationship between gene and species diversity is clarified and ecological patterns of species occurrence emerge. By combining these ecological patterns with the evolutionary patterns inherently revealed by phylogenetic analysis of gene sequence data, we find that it may be possible to understand microbial biodiversity within these systems by using principles similar to those developed by evolutionary ecologists to understand biodiversity of larger species. We hope that such an approach guides microbial ecologists to a more realistic and predictive understanding of microbial species occurrence and responsiveness in both natural and disturbed habitats.
Figures





References
-
- Baas Becking L G M. Geobiologie of inleiding tot de milieukunde. The Hague, The Netherlands: W. P. van Stockum and Zoon N.V.; 1934.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

