The cdr2(+) gene encodes a regulator of G2/M progression and cytokinesis in Schizosaccharomyces pombe
- PMID: 9843577
- PMCID: PMC25645
- DOI: 10.1091/mbc.9.12.3399
The cdr2(+) gene encodes a regulator of G2/M progression and cytokinesis in Schizosaccharomyces pombe
Abstract
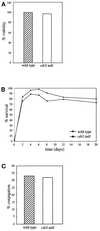
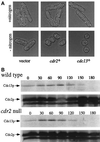
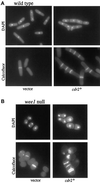
Schizosaccharomyces pombe cells respond to nutrient deprivation by altering G2/M cell size control. The G2/M transition is controlled by activation of the cyclin-dependent kinase Cdc2p. Cdc2p activation is regulated both positively and negatively. cdr2(+) was identified in a screen for regulators of mitotic control during nutrient deprivation. We have cloned cdr2(+) and have found that it encodes a putative serine-threonine protein kinase that is related to Saccharomyces cerevisiae Gin4p and S. pombe Cdr1p/Nim1p. cdr2(+) is not essential for viability, but cells lacking cdr2(+) are elongated relative to wild-type cells, spending a longer period of time in G2. Because of this property, upon nitrogen deprivation cdr2(+) mutants do not arrest in G1, but rather undergo another round of S phase and arrest in G2 from which they are able to enter a state of quiescence. Genetic evidence suggests that cdr2(+) acts as a mitotic inducer, functioning through wee1(+), and is also important for the completion of cytokinesis at 36 degrees C. Defects in cytokinesis are also generated by the overproduction of Cdr2p, but these defects are independent of wee1(+), suggesting that cdr2(+) encodes a second activity involved in cytokinesis.
Figures




References
-
- Aligue R, Wu L, Russell P. Regulation of Schizosaccharomyces pombe Wee1 tyrosine kinase. J Biol Chem. 1997;272:13320–13325. - PubMed
-
- Bähler J, Wu J, Longtine MS, Shah NS, McKenzie A, III, Steever AB, Wach A, Philippsen P, Pringle JR. Heterologous molecules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. - PubMed
-
- Balasubramanian MK, McCollum D, Gould KL. Cytokinesis in the fission yeast Schizosaccharomyes pombe. Methods Enzymol. 1997;283:494–506. - PubMed
-
- Belenguer P, Pelloquin L, Oustrin ML, Ducommun B. Role of the fission yeast nim1 protein kinase in the cell cycle response to nutritional signals. Biochem Biophys Res Commun. 1997;232:204–208. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

