Structural assignments to the Mycoplasma genitalium proteins show extensive gene duplications and domain rearrangements
- PMID: 9843945
- PMCID: PMC24505
- DOI: 10.1073/pnas.95.25.14658
Structural assignments to the Mycoplasma genitalium proteins show extensive gene duplications and domain rearrangements
Abstract
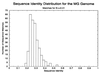
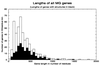
The parasitic bacterium Mycoplasma genitalium has a small, reduced genome with close to a basic set of genes. As a first step toward determining the families of protein domains that form the products of these genes, we have used the multiple sequence programs PSI-BLAST and GEANFAMMER to match the sequences of the 467 gene products of M. genitalium to the sequences of the domains that form proteins of known structure [Protein Data Bank (PDB) sequences]. PDB sequences (274) match all of 106 M. genitalium sequences and some parts of another 85; thus, 41% of its total sequences are matched in all or part. The evolutionary relationships of the PDB domains that match M. genitalium are described in the structural classification of proteins (SCOP) database. Using this information, we show that the domains in the matched M. genitalium sequences come from 114 superfamilies and that 58% of them have arisen by gene duplication. This level of duplication is more than twice that found by using pairwise sequence comparisons. The PDB domain matches also describe the domain structure of the matched sequences: just over a quarter contain one domain and the rest have combinations of two or more domains.
Figures
References
-
- Murzin A G, Brenner S E, Hubbard T, Chothia C. J Mol Biol. 1995;247:536–540. - PubMed
-
- Orengo C A, Michie A D, Jones S, Jones D T, Swindells M B, Thornton J M. Structure (London) 1997;5:1093–1108. - PubMed
-
- Ingram V. Nature (London) 1961;189:704–708. - PubMed
-
- Rossmann M G, Moras D, Olsen K W. Nature (London) 1974;250:194–199. - PubMed
-
- Patthy L. Curr Opin Struct Biol. 1991;1:351–361.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

