The increased level of beta1,4-galactosyltransferase required for lactose biosynthesis is achieved in part by translational control
- PMID: 9843970
- PMCID: PMC24530
- DOI: 10.1073/pnas.95.25.14805
The increased level of beta1,4-galactosyltransferase required for lactose biosynthesis is achieved in part by translational control
Abstract
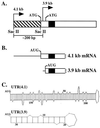
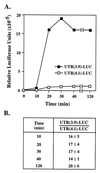
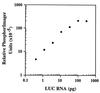
beta1,4-Galactosyltransferase (beta4GalT-I) participates in both glycoconjugate biosynthesis (ubiquitous activity) and lactose biosynthesis (mammary gland-specific activity). In somatic tissues, transcription of the mammalian beta4GalT-I gene results in a 4.1-kb mRNA and a 3.9-kb mRNA as a consequence of initiation at two start sites separated by approximately 200 bp. In the mammary gland, coincident with the increased beta4GalT-I enzyme level ( approximately 50-fold) required for lactose biosynthesis, there is a switch from the 4.1-kb start site to the preferential use of the 3.9-kb start site, which is governed by a stronger tissue-restricted promoter. The use of the 3.9-kb start site results in a beta4GalT-I transcript in which the 5'- untranslated region (UTR) has been truncated from approximately 175 nt to approximately 28 nt. The 5'-UTR of the 4.1-kb transcript [UTR(4.1)] is predicted to contain extensive secondary structure, a feature previously shown to reduce translational efficiency of an mRNA. In contrast, the 5'-UTR of the 3.9-kb mRNA [UTR(3.9)] lacks extensive secondary structure; thus, this transcript is predicted to be more efficiently translated relative to the 4.1-kb mRNA. To test this prediction, constructs were assembled in which the respective 5'-UTRs were fused to the luciferase-coding sequence and enzyme levels were determined after translation in vitro and in vivo. The luciferase mRNA containing the truncated UTR(3.9) was translated more efficiently both in vitro (approximately 14-fold) and in vivo (3- to 5-fold) relative to the luciferase mRNA containing the UTR(4.1). Consequently, in addition to control at the transcriptional level, beta4GalT-I enzyme levels are further augmented in the lactating mammary gland as a result of translational control.
Figures

Similar articles
-
Beta1,4-galactosyltransferase and lactose biosynthesis: recruitment of a housekeeping gene from the nonmammalian vertebrate gene pool for a mammary gland specific function.J Mammary Gland Biol Neoplasia. 1998 Jul;3(3):315-24. doi: 10.1023/a:1018719612087. J Mammary Gland Biol Neoplasia. 1998. PMID: 10819517 Review.
-
Transcriptional regulation of murine beta1,4-galactosyltransferase in somatic cells. Analysis of a gene that serves both a housekeeping and a mammary gland-specific function.J Biol Chem. 1996 Mar 1;271(9):5131-42. doi: 10.1074/jbc.271.9.5131. J Biol Chem. 1996. PMID: 8617793
-
Murine beta 1,4-galactosyltransferase: round spermatid transcripts are characterized by an extended 5'-untranslated region.Glycobiology. 1992 Aug;2(4):361-8. doi: 10.1093/glycob/2.4.361. Glycobiology. 1992. PMID: 1384819
-
Murine beta 1,4-galactosyltransferase: both the amounts and structure of the mRNA are regulated during spermatogenesis.Proc Natl Acad Sci U S A. 1990 Jan;87(2):791-5. doi: 10.1073/pnas.87.2.791. Proc Natl Acad Sci U S A. 1990. PMID: 1689054 Free PMC article.
-
What Is the Impact of mRNA 5' TL Heterogeneity on Translational Start Site Selection and the Mammalian Cellular Phenotype?Front Genet. 2016 Aug 31;7:156. doi: 10.3389/fgene.2016.00156. eCollection 2016. Front Genet. 2016. PMID: 27630668 Free PMC article. Review.
Cited by
-
CART classification of human 5' UTR sequences.Genome Res. 2000 Nov;10(11):1807-16. doi: 10.1101/gr.gr-1460r. Genome Res. 2000. PMID: 11076865 Free PMC article.
-
Traveling for the glycosphingolipid path.Glycoconj J. 2000 Jul-Sep;17(7-9):627-47. doi: 10.1023/a:1011086929064. Glycoconj J. 2000. PMID: 11421354 Review.
-
Analytical tools for the study of cellular glycosylation in the immune system.Front Immunol. 2013 Dec 11;4:451. doi: 10.3389/fimmu.2013.00451. Front Immunol. 2013. PMID: 24376449 Free PMC article. Review.
-
The mammary gland and its origin during synapsid evolution.J Mammary Gland Biol Neoplasia. 2002 Jul;7(3):225-52. doi: 10.1023/a:1022896515287. J Mammary Gland Biol Neoplasia. 2002. PMID: 12751889 Review.
-
Beta1,4-galactosyltransferase and lactose biosynthesis: recruitment of a housekeeping gene from the nonmammalian vertebrate gene pool for a mammary gland specific function.J Mammary Gland Biol Neoplasia. 1998 Jul;3(3):315-24. doi: 10.1023/a:1018719612087. J Mammary Gland Biol Neoplasia. 1998. PMID: 10819517 Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

