Gene dosage and stochastic effects determine the severity and direction of uniparental ribosomal RNA gene silencing (nucleolar dominance) in Arabidopsis allopolyploids
- PMID: 9843986
- PMCID: PMC24546
- DOI: 10.1073/pnas.95.25.14891
Gene dosage and stochastic effects determine the severity and direction of uniparental ribosomal RNA gene silencing (nucleolar dominance) in Arabidopsis allopolyploids
Abstract
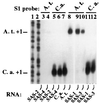
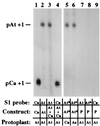
Nucleolar dominance is an epigenetic phenomenon in which one parental set of ribosomal RNA (rRNA) genes is silenced in an interspecific hybrid. In natural Arabidopsis suecica, an allotetraploid (amphidiploid) hybrid of Arabidopsis thaliana and Cardaminopsis arenosa, the A. thaliana rRNA genes are repressed. Interestingly, A. thaliana rRNA gene silencing is variable in synthetic Arabidopsis suecica F1 hybrids. Two generations are needed for A. thaliana rRNA genes to be silenced in all lines, revealing a species-biased direction but stochastic onset to nucleolar dominance. Backcrossing synthetic A. suecica to tetraploid A. thaliana yielded progeny with active A. thaliana rRNA genes and, in some cases, silenced C. arenosa rRNA genes, showing that the direction of dominance can be switched. The hypothesis that naturally dominant rRNA genes have a superior binding affinity for a limiting transcription factor is inconsistent with dominance switching. Inactivation of a species-specific transcription factor is argued against by showing that A. thaliana and C. arenosa rRNA genes can be expressed transiently in the other species. Transfected A. thaliana genes are also active in A. suecica protoplasts in which chromosomal A. thaliana genes are repressed. Collectively, these data suggest that nucleolar dominance is a chromosomal phenomenon that results in coordinate or cooperative silencing of rRNA genes.
Figures





References
-
- Navashin M S. Proc Int Conf Genet Eng. 1928;5:1148–1152.
-
- Navashin M. Cytologia. 1934;5:169–203.
-
- McClintock B. Z Zellforsch Mikrosk Anat. 1934;21:294–328.
-
- Wallace H, Birnstiel M L. Biochem Biophys Acta. 1966;114:296–310. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

