NPH4, a conditional modulator of auxin-dependent differential growth responses in Arabidopsis
- PMID: 9847100
- PMCID: PMC34742
- DOI: 10.1104/pp.118.4.1265
NPH4, a conditional modulator of auxin-dependent differential growth responses in Arabidopsis
Abstract
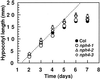
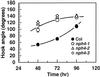
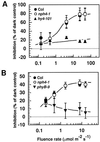
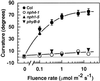
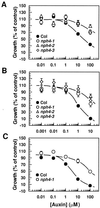
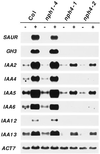
Although sessile in nature, plants are able to use a number of mechanisms to modify their morphology in response to changing environmental conditions. Differential growth is one such mechanism. Despite its importance in plant development, little is known about the molecular events regulating the establishment of differential growth. Here we report analyses of the nph4 (nonphototropic hypocotyl) mutants of Arabidopsis that suggest that the NPH4 protein plays a central role in the modulation of auxin-dependent differential growth. Results from physiological studies demonstrate that NPH4 activity is conditionally required for a number of differential growth responses, including phototropism, gravitropism, phytochrome-dependent hypocotyl curvature, apical hook maintenance, and abaxial/adaxial leaf-blade expansion. The nph4 mutants exhibited auxin resistance and severely impaired auxin-dependent gene expression, indicating that the defects associated with differential growth likely arise because of altered auxin responsiveness. Moreover, the auxin signaling events mediating phototropism are genetically correlated with the abundance of the NPH4 protein.
Figures


References
-
- Abel S, Nguyen MD, Theologis A. The PS-IAA4/5-like family of early auxin inducible messenger RNAs in Arabidopsis thaliana. J Mol Biol. 1995;251:533–549. - PubMed
-
- Ausubel F, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Short Protocols in Molecular Biology. New York: John Wiley & Sons; 1995.
-
- Bell C, Ecker JR. Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics. 1994;19:137–144. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

