Targeting of the visna virus tat protein to AP-1 sites: interactions with the bZIP domains of fos and jun in vitro and in vivo
- PMID: 9847304
- PMCID: PMC103805
- DOI: 10.1128/JVI.73.1.37-45.1999
Targeting of the visna virus tat protein to AP-1 sites: interactions with the bZIP domains of fos and jun in vitro and in vivo
Abstract
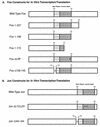
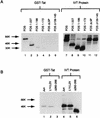
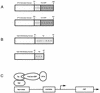
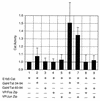
The visna virus Tat protein is required for efficient viral transcription from the visna virus long terminal repeat (LTR). AP-1 sites within the visna virus LTR, which can be bound by the cellular transcription factors Fos and Jun, are also necessary for Tat-mediated transcriptional activation. A potential mechanism by which the visna virus Tat protein could target the viral promoter is by protein-protein interactions with Fos and/or Jun bound to AP-1 sites in the visna virus LTR. Once targeted to the visna virus promoter, the Tat protein could then interact with basal transcription factors to activate transcription. To examine protein-protein interactions with cellular proteins at the visna virus promoter, we used an in vitro protein affinity chromatography assay and electrophoretic mobility shift assay, in addition to an in vivo two-hybrid assay, to show that the visna virus Tat protein specifically interacts with the cellular transcription factors Fos and Jun and the basal transcription factor TBP (TATA binding protein). The Tat domain responsible for interactions with Fos and Jun was localized to an alpha-helical domain within amino acids 34 to 69 of the protein. The TBP binding domain was localized to amino acids 1 to 38 of Tat, a region previously described by our laboratory as the visna virus Tat activation domain. The bZIP domains of Fos and Jun were found to be important for the interactions with Tat. Mutations within the basic domains of Fos and Jun abrogated binding to Tat in the in vitro assays. The visna virus Tat protein was also able to interact with covalently cross-linked Fos and Jun dimers. Thus, the visna virus Tat protein appears to target AP-1 sites in the viral promoter in a mechanism similar to the interaction of human T-cell leukemia virus type 1 Tax with the cellular transcription factor CREB, by binding the basic domains of an intact bZIP dimer. The association between Tat, Fos, and Jun would position Tat proximal to the viral TATA box, where the visna virus Tat activation domain could contact TBP to activate viral transcription.
Figures





Similar articles
-
The leucine domain of the visna virus Tat protein mediates targeting to an AP-1 site in the viral long terminal repeat.J Virol. 1996 Jul;70(7):4338-44. doi: 10.1128/JVI.70.7.4338-4344.1996. J Virol. 1996. PMID: 8676456 Free PMC article.
-
The visna transcriptional activator Tat: effects on the viral LTR and on cellular genes.Virology. 1993 Nov;197(1):236-44. doi: 10.1006/viro.1993.1584. Virology. 1993. PMID: 8212559
-
Molecular mechanisms of visna virus Tat: identification of the targets for transcriptional activation and evidence for a post-transcriptional effect.Virology. 1992 Jun;188(2):438-50. doi: 10.1016/0042-6822(92)90497-d. Virology. 1992. PMID: 1316669
-
Close encounters of many kinds: Fos-Jun interactions that mediate transcription regulatory specificity.Oncogene. 2001 Apr 30;20(19):2438-52. doi: 10.1038/sj.onc.1204385. Oncogene. 2001. PMID: 11402339 Review.
-
fos-jun conspiracy: implications for the cell.Ciba Found Symp. 1990;150:128-37; discussion 137-46. doi: 10.1002/9780470513927.ch9. Ciba Found Symp. 1990. PMID: 2115424 Review.
Cited by
-
Visna virus-induced activation of MAPK is required for virus replication and correlates with virus-induced neuropathology.J Virol. 2002 Jan;76(2):817-28. doi: 10.1128/jvi.76.2.817-828.2002. J Virol. 2002. PMID: 11752171 Free PMC article.
-
Genomic characterization of a slow/low maedi visna virus.Virus Genes. 2004 Oct;29(2):199-210. doi: 10.1023/B:VIRU.0000036380.01957.37. Virus Genes. 2004. PMID: 15284480
-
Human immunodeficiency virus and AIDS: insights from animal lentiviruses.J Virol. 2000 Aug;74(16):7187-95. doi: 10.1128/jvi.74.16.7187-7195.2000. J Virol. 2000. PMID: 10906172 Free PMC article. Review. No abstract available.
-
Expanding possibilities for intervention against small ruminant lentiviruses through genetic marker-assisted selective breeding.Viruses. 2013 Jun 14;5(6):1466-99. doi: 10.3390/v5061466. Viruses. 2013. PMID: 23771240 Free PMC article. Review.
-
Construction and in vitro characterization of attenuated feline immunodeficiency virus long terminal repeat mutant viruses.J Virol. 2001 Jan;75(2):1054-60. doi: 10.1128/JVI.75.2.1054-1060.2001. J Virol. 2001. PMID: 11134320 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

