Colocalization of baculovirus IE-1 and two DNA-binding proteins, DBP and LEF-3, to viral replication factories
- PMID: 9847313
- PMCID: PMC103814
- DOI: 10.1128/JVI.73.1.110-119.1999
Colocalization of baculovirus IE-1 and two DNA-binding proteins, DBP and LEF-3, to viral replication factories
Abstract
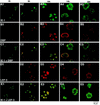
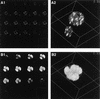
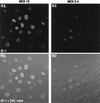
We have recently identified a DNA-binding protein (DBP) from the baculovirus Bombyx mori nucleopolyhedrovirus (BmNPV) which can destabilize double-stranded DNA (V. S. Mikhailov, A. L. Mikhailova, M. Iwanaga, S. Gomi, and S. Maeda, J. Virol. 72:3107-3116, 1998). DBP was found to be an early gene product that was not present in budded or occlusion-derived virions. In order to characterize the localization of DBP during viral replication, BmNPV-infected BmN cells were examined by immunostaining and confocal microscopy with DBP antibodies. DBP first appeared as diffuse nuclear staining at 4 to 6 h postinfection (p.i.) and then localized to several specific foci within the nucleus at 6 to 8 h p.i. After the onset of viral DNA replication at around 8 h p.i., these foci began to enlarge and eventually occupied more than half of the nucleus by 14 h p.i. After the termination of viral DNA replication at about 20 h p.i., the DBP-stained regions appeared to break down into approximately 100 small foci within the nucleus. At 8 h p.i., the distribution of DBP as well as that of IE-1 or LEF-3 (two proteins involved in baculovirus DNA replication) overlapped well with that of DNA replication sites labeled with bromodeoxyuridine incorporation. Double-staining experiments with IE-1 and DBP or IE-1 and LEF-3 further confirmed that, between 8 and 14 h p.i., the distribution of IE-1 and LEF-3 overlapped with that of DBP. However, IE-1 localized to the specific foci prior to DBP or LEF-3 at 4 h p.i. In the presence of aphidicolin, an inhibitor of DNA synthesis, immature foci containing IE-1, LEF-3, and DBP were observed by 8 h p.i. However, the subsequent enlargement of these foci was completely suppressed, suggesting that the enlargement depended upon viral DNA replication. At 4 h p.i., the number of IE-1 foci correlated with the multiplicity of infection (MOI) between 0.4 and 10. At higher MOIs (e.g., 50), the number of foci plateaued at around 15. These results suggested that there are about 15 preexisting sites per nucleus which are associated with the initiation of viral DNA replication and assembly of viral DNA replication factories.
Figures






References
-
- Ahrens C H, Carlson C, Rohrmann G F. Identification, sequence, and transcriptional analysis of lef-3, a gene essential for Orgyia pseudotsugata baculovirus DNA replication. Virology. 1995;210:372–382. - PubMed
-
- Ahrens C H, Rohrmann G F. The DNA polymerase and helicase genes of a baculovirus of Orgyia pseudotsugata. J Gen Virol. 1996;77:825–837. - PubMed
-
- Ahrens C H, Rohrmann G F. Identification of essential trans-acting regions required for DNA replication of the Orgyia pseudotsugata multinucleocapsid nuclear polyhedrosis virus: lef-1 is an essential replication gene. Virology. 1995;207:417–428. - PubMed
-
- Ahrens C H, Rohrmann G F. Replication of Orgyia pseudotsugata baculovirus DNA: lef-2 and ie-1 are essential and ie-2, p34, and Op-iap are stimulatory genes. Virology. 1995;212:650–662. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

