Porcine reproductive and respiratory syndrome virus comparison: divergent evolution on two continents
- PMID: 9847330
- PMCID: PMC103831
- DOI: 10.1128/JVI.73.1.270-280.1999
Porcine reproductive and respiratory syndrome virus comparison: divergent evolution on two continents
Abstract
Porcine reproductive and respiratory syndrome virus (PRRSV) is a recently described arterivirus responsible for disease in swine worldwide. Comparative sequence analysis of 3'-terminal structural genes of the single-stranded RNA viral genome revealed the presence of two genotypic classes of PRRSV, represented by the prototype North American and European strains, VR-2332 and Lelystad virus (LV), respectively. To better understand the evolution and pathogenicity of PRRSV, we obtained the 12,066-base 5'-terminal nucleotide sequence of VR-2332, encoding the viral replication activities, and compared it to those of LV and other arteriviruses. VR-2332 and LV differ markedly in the 5' leader and sections of the open reading frame (ORF) 1a region. The ORF 1b sequence was nearly colinear but varied in similarity of proteins encoded in identified regions. Furthermore, molecular and biochemical analysis of subgenomic mRNA (sgmRNA) processing revealed extensive variation in the number of sgmRNAs which may be generated during infection and in the lengths of noncoding sequence between leader-body junctions and the translation-initiating codon AUG. In addition, VR-2332 and LV select different leader-body junction sites from a pool of similar candidate sites to produce sgmRNA 7, encoding the viral nucleocapsid protein. The presence of substantial variations across the entire genome and in sgmRNA processing indicates that PRRSV has evolved independently on separate continents. The near-simultaneous global emergence of a new swine disease caused by divergently evolved viruses suggests that changes in swine husbandry and management may have contributed to the emergence of PRRS.
Figures


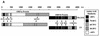
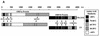
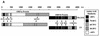
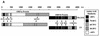
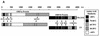
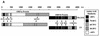


References
-
- Baarsch M J, Wannemuehler M J, Molitor T W, Murtaugh M P. Detection of tumor necrosis factor alpha from porcine alveolar macrophages using an L929 fibroblast bioassay. J Immunol Methods. 1991;140:15–22. - PubMed
-
- Bautista E M, Meulenberg J J, Choi C S, Molitor T W. Structural polypeptides of the American (VR-2332) strain of porcine reproductive and respiratory syndrome virus. Arch Virol. 1996;141:1357–1365. - PubMed
-
- Benfield D A, Nelson E, Collins J E, Harris L, Goyal S M, Robison D, Christianson W T, Morrison R B, Gorcyca D E, Chladek D W. Characterization of swine infertility and respiratory syndrome (SIRS) virus (isolate ATCC VR-2332) J Vet Diagn Investig. 1992;4:127–133. - PubMed
-
- Brinton M A, Gavin E I, Fernandez A V. Genotypic variation among six isolates of lactate dehydrogenase-elevating virus. J Gen Virol. 1986;67:2673–2684. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

