Overlapping signals for transcription and replication at the 3' terminus of the vesicular stomatitis virus genome
- PMID: 9847350
- PMCID: PMC103851
- DOI: 10.1128/JVI.73.1.444-452.1999
Overlapping signals for transcription and replication at the 3' terminus of the vesicular stomatitis virus genome
Abstract
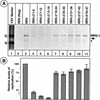
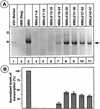
Transcription and replication signals within the negative-sense genomic RNA of vesicular stomatitis virus (VSV) are located at the 3' terminus. To identify these signals, we have used a transcription- and replication-competent minigenome of VSV to generate a series of deletions spanning the first 47 nucleotides at the 3' terminus of the VSV genome corresponding to the leader gene. Analysis of these mutants for their ability to replicate showed that deletion of sequences within the first 24 nucleotides abrogated or greatly reduced the level of replication. Deletion of downstream sequences from nucleotides 25 to 47 reduced the level of replication only to 55 to 70% of that of the parental template. When transcription activity of these templates was measured, the first 24 nucleotides were also found to be required for transcription, since deletion of these sequences blocked or significantly reduced transcription. Downstream sequences from nucleotides 25 to 47 were necessary for optimal levels of transcription. Furthermore, replacement of sequences within the 25 to 47 nucleotides with random heterologous nonviral sequences generated mutant templates that replicated well (65 to 70% of the wild-type levels) but were transcribed poorly (10 to 15% of the wild-type levels). These results suggest that the minimal promoter for transcription and replication could be as small as the first 19 nucleotides and is contained within the 3'-terminal 24 nucleotides of the VSV genome. The sequences from nucleotides 25 to 47 may play a more important role in optimal transcription than in replication. Our results also show that deletion of sequences within the leader gene does not influence the site of transcription reinitiation of the downstream gene.
Figures





Similar articles
-
Replication signals in the genome of vesicular stomatitis virus and its defective interfering particles: identification of a sequence element that enhances DI RNA replication.Virology. 1997 Jun 9;232(2):248-59. doi: 10.1006/viro.1997.8571. Virology. 1997. PMID: 9191838
-
The termini of VSV DI particle RNAs are sufficient to signal RNA encapsidation, replication, and budding to generate infectious particles.Virology. 1995 Jan 10;206(1):760-4. doi: 10.1016/s0042-6822(95)80005-0. Virology. 1995. PMID: 7831839 Free PMC article.
-
Regulation of RNA synthesis by the genomic termini of vesicular stomatitis virus: identification of distinct sequences essential for transcription but not replication.J Virol. 1999 Jan;73(1):297-306. doi: 10.1128/JVI.73.1.297-306.1999. J Virol. 1999. PMID: 9847333 Free PMC article.
-
The 5' terminal trailer region of vesicular stomatitis virus contains a position-dependent cis-acting signal for assembly of RNA into infectious particles.J Virol. 1999 Jan;73(1):307-15. doi: 10.1128/JVI.73.1.307-315.1999. J Virol. 1999. PMID: 9847334 Free PMC article.
-
Basic amino acid residues at the carboxy-terminal eleven amino acid region of the phosphoprotein (P) are required for transcription but not for replication of vesicular stomatitis virus genome RNA.Virology. 1997 Nov 10;238(1):103-14. doi: 10.1006/viro.1997.8823. Virology. 1997. PMID: 9375014
Cited by
-
The family Rhabdoviridae: mono- and bipartite negative-sense RNA viruses with diverse genome organization and common evolutionary origins.Virus Res. 2017 Jan 2;227:158-170. doi: 10.1016/j.virusres.2016.10.010. Epub 2016 Oct 20. Virus Res. 2017. PMID: 27773769 Free PMC article. Review.
-
Transcription and replication initiate at separate sites on the vesicular stomatitis virus genome.Proc Natl Acad Sci U S A. 2002 Jul 9;99(14):9178-83. doi: 10.1073/pnas.152155599. Epub 2002 Jun 27. Proc Natl Acad Sci U S A. 2002. PMID: 12089339 Free PMC article.
-
Vesicular stomatitis viruses resistant to the methylase inhibitor sinefungin upregulate RNA synthesis and reveal mutations that affect mRNA cap methylation.J Virol. 2007 Apr;81(8):4104-15. doi: 10.1128/JVI.02681-06. Epub 2007 Feb 14. J Virol. 2007. PMID: 17301155 Free PMC article.
-
RNA Synthesis and Capping by Non-segmented Negative Strand RNA Viral Polymerases: Lessons From a Prototypic Virus.Front Microbiol. 2019 Jul 10;10:1490. doi: 10.3389/fmicb.2019.01490. eCollection 2019. Front Microbiol. 2019. PMID: 31354644 Free PMC article. Review.
-
How does the polymerase of non-segmented negative strand RNA viruses commit to transcription or genome replication?J Virol. 2024 Aug 20;98(8):e0033224. doi: 10.1128/jvi.00332-24. Epub 2024 Jul 30. J Virol. 2024. PMID: 39078194 Free PMC article. Review.
References
-
- Ausubel F, Brent R, Kingston E, Moore D, Seidman J, Smith J, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley and Sons, Inc.; 1988.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

