The genome of Melanoplus sanguinipes entomopoxvirus
- PMID: 9847359
- PMCID: PMC103860
- DOI: 10.1128/JVI.73.1.533-552.1999
The genome of Melanoplus sanguinipes entomopoxvirus
Abstract
The family Poxviridae contains two subfamilies: the Entomopoxvirinae (poxviruses of insects) and the Chordopoxvirinae (poxviruses of vertebrates). Here we present the first characterization of the genome of an entomopoxvirus (EPV) which infects the North American migratory grasshopper Melanoplus sanguinipes and other important orthopteran pests. The 236-kbp M. sanguinipes EPV (MsEPV) genome consists of a central coding region bounded by 7-kbp inverted terminal repeats and contains 267 open reading frames (ORFs), of which 107 exhibit similarity to previously described genes. The presence of genes not previously described in poxviruses, and in some cases in any other known virus, suggests significant viral adaptation to the arthropod host and the external environment. Genes predicting interactions with host cellular mechanisms include homologues of the inhibitor of apoptosis protein, stress response protein phosphatase 2C, extracellular matrixin metalloproteases, ubiquitin, calcium binding EF-hand protein, glycosyltransferase, and a triacylglyceride lipase. MsEPV genes with putative functions in prevention and repair of DNA damage include a complete base excision repair pathway (uracil DNA glycosylase, AP endonuclease, DNA polymerase beta, and an NAD+-dependent DNA ligase), a photoreactivation repair pathway (cyclobutane pyrimidine dimer photolyase), a LINE-type reverse transcriptase, and a mutT homologue. The presence of these specific repair pathways may represent viral adaptation for repair of environmentally induced DNA damage. The absence of previously described poxvirus enzymes involved in nucleotide metabolism and the presence of a novel thymidylate synthase homologue suggest that MsEPV is heavily reliant on host cell nucleotide pools and the de novo nucleotide biosynthesis pathway. MsEPV and lepidopteran genus B EPVs lack genome colinearity and exhibit a low level of amino acid identity among homologous genes (20 to 59%), perhaps reflecting a significant evolutionary distance between lepidopteran and orthopteran viruses. Divergence between MsEPV and the Chordopoxvirinae is indicated by the presence of only 49 identifiable chordopoxvirus homologues, low-level amino acid identity among these genes (20 to 48%), and the presence in MsEPV of 43 novel ORFs in five gene families. Genes common to both poxvirus subfamilies, which include those encoding enzymes involved in RNA transcription and modification, DNA replication, protein processing, virion assembly, and virion structural proteins, define the genetic core of the Poxviridae.
Figures

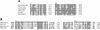
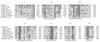
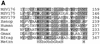
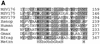




References
-
- Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Andres G, Simon-Mateo C, Vinuela E. Characterization of two African swine fever virus 220-kDa proteins: a precursor of the major structural protein p150 and an oligomer of phosphoprotein p32. Virology. 1993;194:284–293. - PubMed
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

