Demonstration that orf2 encodes the feline immunodeficiency virus transactivating (Tat) protein and characterization of a unique gene product with partial rev activity
- PMID: 9847366
- PMCID: PMC103867
- DOI: 10.1128/JVI.73.1.608-617.1999
Demonstration that orf2 encodes the feline immunodeficiency virus transactivating (Tat) protein and characterization of a unique gene product with partial rev activity
Abstract
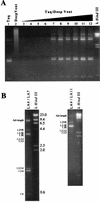
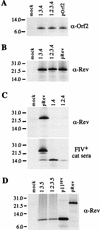
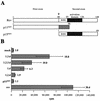
The long PCR technique was used to amplify the three size classes of viral mRNAs produced in cells infected by feline immunodeficiency virus (FIV). We identified in the env region a new splice acceptor site that generated two transcripts, each coding for an 11-kDa protein, p11(rev), whose function is unknown. The small-size class of mRNAs included two bicistronic orf2/rev mRNAs and two rev-like mRNAs, consisting only of the second exon of rev and coding for a 15-kDa protein, p15(rev). p15(rev) contained the minimal effector domain of Rev and was sufficient to mediate partial Rev activity. The bicistronic mRNAs encoded two distinct proteins, one of 23 kDa corresponding to Rev and a 9-kDa protein encoded by the orf2 gene. The orf2 gene product is a protein of 79 amino acids with characteristics similar to those of the Tat (transactivator) proteins of the ungulate lentiviruses. Transient expression assays, using the FIV long terminal repeat (LTR) to drive transcription of the bacterial gene for chloramphenicol acetyltransferase demonstrated that the orf2 gene transactivates gene expression an average of 14- to 20-fold above the basal level. Deletion mutants of the FIV LTR were generated to locate sequences responsive to transactivation mediated by the orf2 gene. A 5' deletion mutant that removed the AP1 site resulted in residual low-level transactivation by orf2. Further experiments using LTR mutants with internal deletions identified three regions located between positions -126 and -47 relative to the cap site that were important for orf2-directed transactivation. These regions include the AP1 site, a C/EBP tandem repeat, and an ATF site.
Figures



References
-
- Ali S A, Steinkasserer A. PCR-ligation-PCR mutagenesis: a protocol for creating gene fusions and mutations. BioTechniques. 1995;18:746–750. - PubMed
-
- Chatterji, U., and J. H. Elder. Unpublished observation.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

