The essential Gcd10p-Gcd14p nuclear complex is required for 1-methyladenosine modification and maturation of initiator methionyl-tRNA
- PMID: 9851972
- PMCID: PMC317256
- DOI: 10.1101/gad.12.23.3650
The essential Gcd10p-Gcd14p nuclear complex is required for 1-methyladenosine modification and maturation of initiator methionyl-tRNA
Abstract
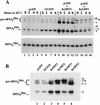
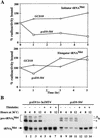
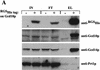
Gcd10p and Gcd14p are essential proteins required for the initiation of protein synthesis and translational repression of GCN4 mRNA. The phenotypes of gcd10 mutants were suppressed by high-copy-number IMT genes, encoding initiator methionyl tRNA (tRNAiMet), or LHP1, encoding the yeast homolog of the human La autoantigen. The gcd10-504 mutation led to a reduction in steady-state levels of mature tRNAiMet, attributable to increased turnover rather than decreased synthesis of pre-tRNAiMet. Remarkably, the lethality of a GCD10 deletion was suppressed by high-copy-number IMT4, indicating that its role in expression of mature tRNAiMet is the essential function of Gcd10p. A gcd14-2 mutant also showed reduced amounts of mature tRNAiMet, but in addition, displayed a defect in pre-tRNAiMet processing. Gcd10p and Gcd14p were found to be subunits of a protein complex with prominent nuclear localization, suggesting a direct role in tRNAiMet maturation. The chromatographic behavior of elongator and initiator tRNAMet on a RPC-5 column indicated that both species are altered structurally in gcd10Delta cells, and analysis of base modifications revealed that 1-methyladenosine (m1A) is undetectable in gcd10Delta tRNA. Interestingly, gcd10 and gcd14 mutations had no effect on processing or accumulation of elongator tRNAMet, which also contains m1A at position 58, suggesting a unique requirement for this base modification in initiator maturation.
Figures








References
-
- Asano K, Phan L, Anderson J, Hinnebusch AG. Complex formation by all five homologues of mammalian translation initiation factor 3 subunits from yeast Saccharomyces cerevisiae. J Biol Chem. 1998;273:18573–18585. - PubMed
-
- Boeke JD, Trueheart J, Natsoulis G, Fink GR. 5-Fluoroorotic acid as a selective agent in yeast molecular genes. Methods Enzymol. 1987;154:164–175. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
