Ligand-induced cleavage and regulation of nuclear entry of Notch in Drosophila melanogaster embryos
- PMID: 9851979
- PMCID: PMC317253
- DOI: 10.1101/gad.12.23.3728
Ligand-induced cleavage and regulation of nuclear entry of Notch in Drosophila melanogaster embryos
Abstract
Notch, a transmembrane protein found in a wide range of organisms, is a component of a pathway that mediates cell-fate decisions that involve intercellular communication. In this paper, we show that in Drosophila melanogaster, Notch (N) is processed in a ligand-dependent fashion to generate phosphorylated, soluble intracellular derivatives. Suppressor of Hairless [Su(H)] is predominantly associated with soluble intracellular N. It has been demonstrated by others that N has access to the nucleus, and we show that when tethered directly to DNA, the cytoplasmic domain of N can activate transcription. Conversely, a viral activator fused to Su(H) can substitute for at least some N functions during embryogenesis. We suggest that one function of soluble forms of N is to bind to Su(H), and in the nucleus, to act directly as a transcriptional transactivator of the latter protein. Although N has functional nuclear localization signals, the N/Su(H) complex accumulates in the cytoplasm and on membranes suggesting that its nuclear entry is regulated. Localization studies in cultured cells and embryos suggest that Su(H) plays a role in this regulation, with the relative levels of Delta, N and Su(H) determining whether a N/Su(H) complex enters the nucleus.
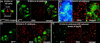
Figures









References
-
- Artavanis-Tsakonas S, Matsuno K, Fortini ME. Notch signaling. Science. 1995;268:225–232. - PubMed
-
- Azpiazu N, Frasch M. Tinman and bagpipe: Two homeo box genes that determine cell fates in the dorsal mesoderm of Drosophila. Genes & Dev. 1993;7:1325–1340. - PubMed
-
- Bailey AM, Posakony JW. Suppressor of Hairless directly activates transcription of enhancer of split complex genes in response to Notch receptor activity. Genes & Dev. 1995;9:2609–2622. - PubMed
-
- Blaumueller CM, Qi H, Zagouras P, Artavanis-Tsakonas S. Intracellular cleavage of Notch leads to a heterodimeric receptor on the plasma membrane. Cell. 1997;90:281–291. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
