Physiological role for the GlnK protein of enteric bacteria: relief of NifL inhibition under nitrogen-limiting conditions
- PMID: 9852012
- PMCID: PMC107771
- DOI: 10.1128/JB.180.24.6661-6667.1998
Physiological role for the GlnK protein of enteric bacteria: relief of NifL inhibition under nitrogen-limiting conditions
Abstract
In Klebsiella pneumoniae, NifA-dependent transcription of nitrogen fixation (nif) genes is inhibited by a flavoprotein, NifL, in the presence of molecular oxygen and/or combined nitrogen. We recently demonstrated that the general nitrogen regulator NtrC is required to relieve NifL inhibition under nitrogen (N)-limiting conditions. We provide evidence that the sole basis for the NtrC requirement is its role as an activator of transcription for glnK, which encodes a PII-like allosteric effector. Relief of NifL inhibition is a unique physiological function for GlnK in that the structurally related GlnB protein of enteric bacteria-apparently a paralogue of GlnK-cannot substitute. Unexpectedly, although covalent modification of GlnK by uridylylation normally occurs under N-limiting conditions, several lines of evidence indicate that uridylylation is not required for relief of NifL inhibition. When GlnK was synthesized constitutively from non-NtrC-dependent promoters, it was able to relieve NifL inhibition in the absence of uridylyltransferase, the product of the glnD gene, and under N excess conditions. Moreover, an altered form of GlnK, GlnKY51N, which cannot be uridylylated due to the absence of the requisite tyrosine, was still able to relieve NifL inhibition.
Figures
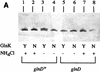

Similar articles
-
Studies on the roles of GlnK and GlnB in regulating Klebsiella pneumoniae NifL-dependent nitrogen control.FEMS Microbiol Lett. 1999 Nov 15;180(2):263-70. doi: 10.1111/j.1574-6968.1999.tb08805.x. FEMS Microbiol Lett. 1999. PMID: 10556721
-
Role of Escherichia coli nitrogen regulatory genes in the nitrogen response of the Azotobacter vinelandii NifL-NifA complex.J Bacteriol. 2001 May;183(10):3076-82. doi: 10.1128/JB.183.10.3076-3082.2001. J Bacteriol. 2001. PMID: 11325935 Free PMC article.
-
NtrC is required for control of Klebsiella pneumoniae NifL activity.J Bacteriol. 1997 Dec;179(23):7446-55. doi: 10.1128/jb.179.23.7446-7455.1997. J Bacteriol. 1997. PMID: 9393710 Free PMC article.
-
Regulation of nitrogen fixation in Klebsiella pneumoniae and Azotobacter vinelandii: NifL, transducing two environmental signals to the nif transcriptional activator NifA.J Mol Microbiol Biotechnol. 2002 May;4(3):235-42. J Mol Microbiol Biotechnol. 2002. PMID: 11931553 Review.
-
Azospirillum, a free-living nitrogen-fixing bacterium closely associated with grasses: genetic, biochemical and ecological aspects.FEMS Microbiol Rev. 2000 Oct;24(4):487-506. doi: 10.1111/j.1574-6976.2000.tb00552.x. FEMS Microbiol Rev. 2000. PMID: 10978548 Review.
Cited by
-
Signal transduction protein P(II) is required for NtcA-regulated gene expression during nitrogen deprivation in the cyanobacterium Synechococcus elongatus strain PCC 7942.J Bacteriol. 2003 Apr;185(8):2582-91. doi: 10.1128/JB.185.8.2582-2591.2003. J Bacteriol. 2003. PMID: 12670983 Free PMC article.
-
Mutagenesis and functional characterization of the glnB, glnA, and nifA genes from the photosynthetic bacterium Rhodospirillum rubrum.J Bacteriol. 2000 Feb;182(4):983-92. doi: 10.1128/JB.182.4.983-992.2000. J Bacteriol. 2000. PMID: 10648524 Free PMC article.
-
Effect of P(II) and its homolog GlnK on reversible ADP-ribosylation of dinitrogenase reductase by heterologous expression of the Rhodospirillum rubrum dinitrogenase reductase ADP-ribosyl transferase-dinitrogenase reductase-activating glycohydrolase regulatory system in Klebsiella pneumoniae.J Bacteriol. 2001 Mar;183(5):1610-20. doi: 10.1128/JB.183.5.1610-1620.2001. J Bacteriol. 2001. PMID: 11160092 Free PMC article.
-
Characterization of GlnK1 from Methanosarcina mazei strain Gö1: complementation of an Escherichia coli glnK mutant strain by GlnK1.J Bacteriol. 2002 Feb;184(4):1028-40. doi: 10.1128/jb.184.4.1028-1040.2002. J Bacteriol. 2002. PMID: 11807063 Free PMC article.
-
Resource Allocation During the Transition to Diazotrophy in Klebsiella oxytoca.Front Microbiol. 2021 Aug 9;12:718487. doi: 10.3389/fmicb.2021.718487. eCollection 2021. Front Microbiol. 2021. PMID: 34434180 Free PMC article.
References
-
- Adler S P, Purich D, Stadtman E. Cascade control of Escherichia coli glutamine synthetase. Properties of the PII regulatory protein and the uridylyltransferase-uridylyl-removing enzyme. J Biol Chem. 1975;250:6264–6272. - PubMed
-
- Allikmets R, Gerrard B, Court D, Dean M. Cloning and organization of the abc and mdl genes of Escherichia coli: relationship to eukaryotic multidrug resistance. Gene. 1993;136:231–236. - PubMed
-
- Arkinson, M. R., and A. J. Ninfa. Unpublished data.
-
- Atkinson M R, Ninfa A J. Role of the GlnK signal transduction protein in the regulation of nitrogen assimilation in Escherichia coli. Mol Microbiol. 1998;29:431–448. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

